Structural and Biological Characterisation of the Gut-Associated Cyclophilin B Isoforms from Caenorhabditis Elegans
Picken, N.C., Eschenlauer, S., Taylor, P., Page, A.P., Walkinshaw, M.D.(2002) J Mol Biol 322: 15
- PubMed: 12215411
- DOI: https://doi.org/10.1016/s0022-2836(02)00712-x
- Primary Citation of Related Structures:
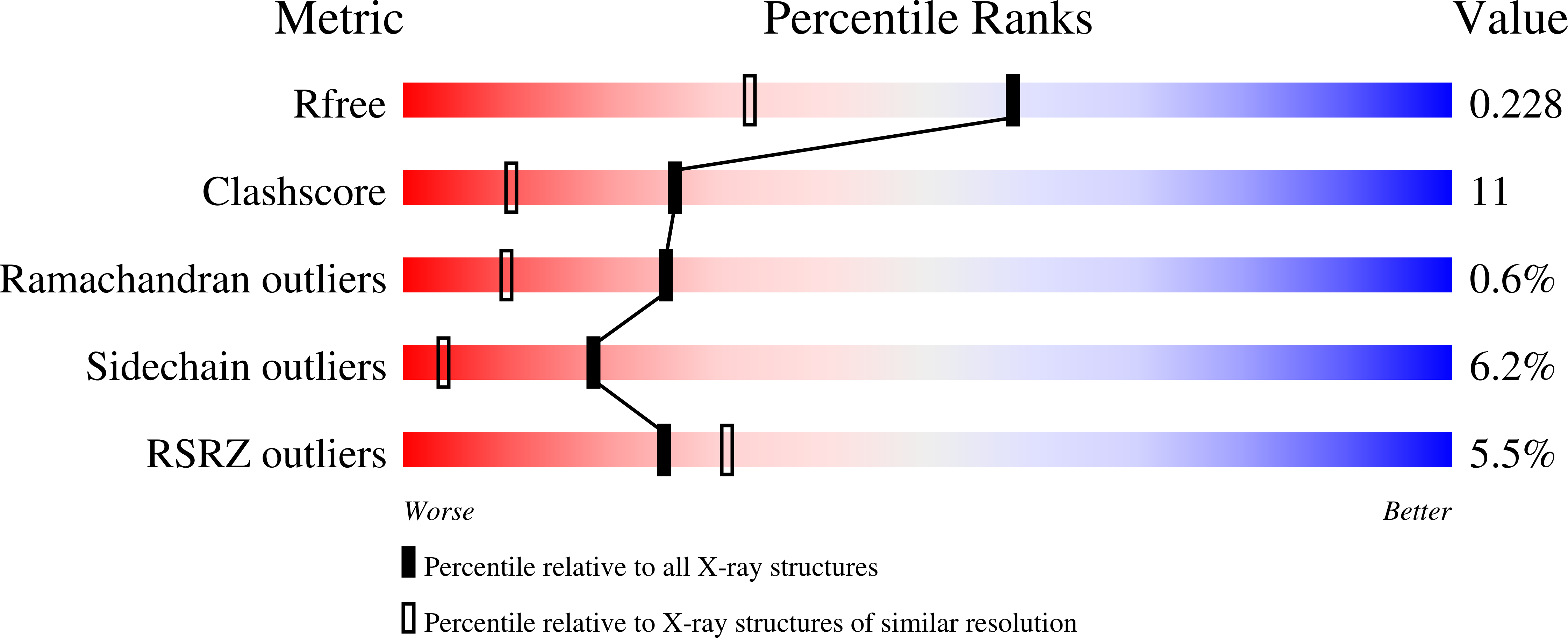
1H0P - PubMed Abstract:
The free-living nematode Caenorhabditis elegans expresses 18 cyclophilin isoforms, eight of which are conserved single domain forms, comprising two closely related secreted or type B forms (CYP-5 and CYP-6). Recombinant CYP-5 has been purified, crystallised and the X-ray structure solved to a resolution of 1.75A. The detailed molecular architecture most strongly resembles the structure of human cyclophilin B with conserved changes in loop structure and N and C-terminal extensions. Interestingly, the active site pocket is occupied by a molecule of dithiothreitol though this has little effect on the geometry of the active site which is similar to other cyclophilin structures. The peptidyl-prolyl isomerase activity of CYP-5 has been characterised against the substrate N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide, and gives a k(cat)/K(m) value of 3.6x10(6)M(-1)s(-1) that compares with a value of 6.3x10(6)M(-1)s(-1) for human cyclophilin B. The immunosuppressive drug cyclosporin A binds and inhibits CYP-5 with an IC(50) value of 50nM, which is comparable to the value of 84nM found for human cyclophilin B. CYP-6 has 67% sequence identity with CYP-5 and a molecular model was built based on the CYP-5 crystal structure. The model shows that CYP-5 and CYP-6 are likely to have very similar structures, but with a markedly increased number of negative charges distributed around the surface of CYP-6. The spatial expression patterns of the cyclophilin B isoforms were examined using transgenic animals carrying a LacZ reporter fusion to these genes, and both cyp-5 and cyp-6 are found to be expressed in an overlapping fashion in the nematode gut. The temporal expression pattern of cyp-5 was further determined and revealed a constitutive expression pattern, with highest abundance levels being found in the embryo.
Organizational Affiliation:
Structural Biochemistry Group, Institute of Cell and Molecular Biology, University of Edinburgh, Michael Swann Building, UK.