Structural variation among retroviral primer-DNA junctions: solution structure of the HIV-1 (-)-strand Okazaki fragment r(gcca)d(CTGC).d(GCAGTGGC).
Fedoroff, O.Y.u., Salazar, M., Reid, B.R.(1996) Biochemistry 35: 11070-11080
- PubMed: 8780509
- DOI: https://doi.org/10.1021/bi9607822
- Primary Citation of Related Structures:
1GTC - PubMed Abstract:
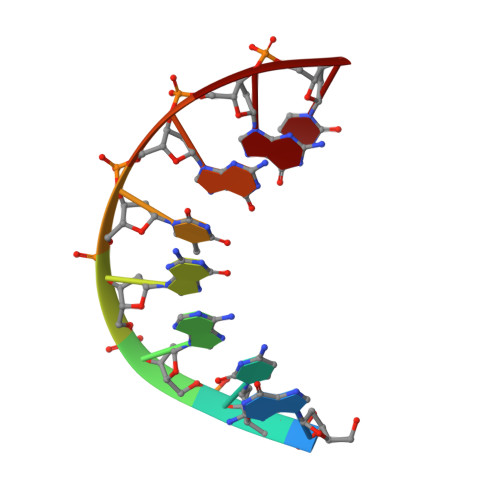
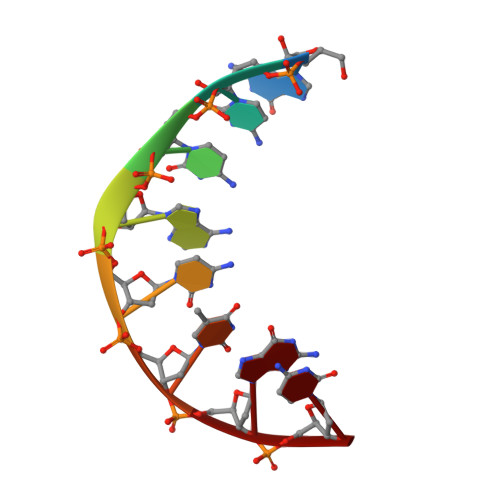
The three-dimensional solution structure of the hybrid-chimeric duplex r(gcca)d(CTGC).d(GCAGTGGC) has been determined by two-dimensional NMR, restrained molecular dynamics (rMD), and NOE back-calculation methods. This chimera, consisting of a chimeric RNA-DNA strand and its complementary DNA strand, is formed after priming (-)-strand DNA synthesis by tRNA(Lys3) and subsequent (+)-strand DNA synthesis by reverse transcriptase and is an obligatory intermediate in the formation of double-stranded DNA prior to HIV-1 retrovirus integration. The duplex consists of two different types of double helix: a hybrid form (H-form) and a B-form structure connected by a junction. It is chemically similar to several other Okazaki fragments whose structures have been previously determined in our laboratory. However, some structural parameters are not the same and were found to be sequence dependent. In particular, the sugar conformations at the DNA base pair proximal to the hybrid segment vary from O4'-endo to C2'-endo depending on the base composition. The position of the transition from the relatively wide groove of H-form to the narrow groove of B-form is also sequence dependent, occurring either exactly at the RNA-DNA junction or within the purely DNA segment of the chimera-as is the case in the structure of the present HIV-1 (-)-strand primer. This structural change produces a kink at the DNA-DNA step adjacent to the RNA-DNA junction in the HIV-1 (-)-strand primer. The sequence dependence of structures of RNA-DNA chimeric duplexes may be responsible for the variable cleavage pattern of different Okazaki fragments by reverse transcriptase RNase H.
Organizational Affiliation:
Chemistry Department, University of Washington, Seattle 98195, USA.