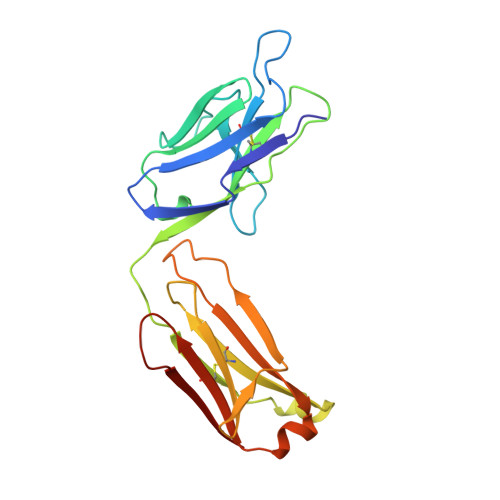
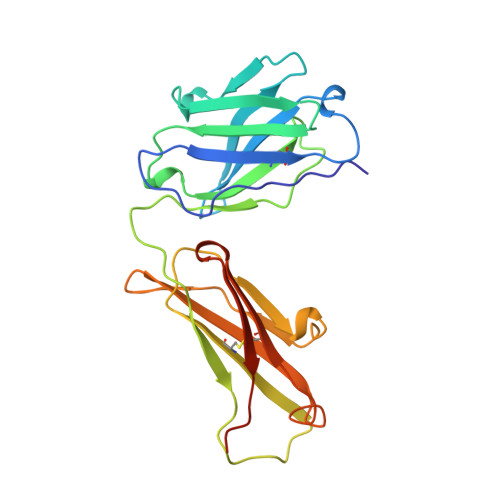
The rational construction of an antibody against cystatin: lessons from the crystal structure of an artificial Fab fragment.
Schiweck, W., Skerra, A.(1997) J Mol Biol 268: 934-951
- PubMed: 9180382
- DOI: https://doi.org/10.1006/jmbi.1997.1006
- Primary Citation of Related Structures:
1GPO - PubMed Abstract:
In a protein design study the artificial antibody M41 was modelled with its binding site complementary to the protease inhibitor cystatin, which was chosen as a structurally well-characterized "antigen". The modelling of M41 took advantage of the crystal structure of the anti-lysozyme antibody HyHEL-10 as a structural template. Its combining site was reshaped by replacing 19 amino acid side-chains in the hypervariable loops. In addition, ten amino acid residues were substituted in the framework regions. The crystal structure of the corresponding antibody model M41, which was produced as an F(ab) fragment in Escherichia coli, was determined at a resolution of 1.95 A. The crystals exhibited symmetry of the space group P2(1)2(1)2(1) (a = 96.5 A; b = 103.5 A; c = 113.6 A) with two F(ab) fragments in the asymmetric unit, which were independently refined (final R-factor 21.7%). The resulting coordinates were used for a detailed comparison with the modelled protein structure. It was found that the mutual arrangement of the six complementarity-determining regions as well as most of their backbone conformation had been correctly predicted. One major difference that was detected for the conformation of a five residue insertion in complementarity-determining region L1 could be explained by an erroneously defined segment in the structure of the antibody 4-4-20, which had been used as a template for this loop. In the light of more recent crystallographic data it appears that this segment adopts a new canonical structure. Apart from this region, most of the side-chains in the antigen-binding site had been properly placed in the M41 model. There was however one important exception concerning Trp H98, whose side-chain conformation had been kept as it appeared in HyHEL-10. The differing orientation of this residue in the model compared with the crystal structure of the artificial F(ab) fragment M41 explains why an antigen affinity could not be detected so far. The detailed analysis of this and other, more subtle deviations suggests how to make this F(ab) fragment function by introducing a few additional amino acid changes into M41.
Organizational Affiliation:
Max-Planck-Institut für Biophysik, Frankfurt am Main, Germany.