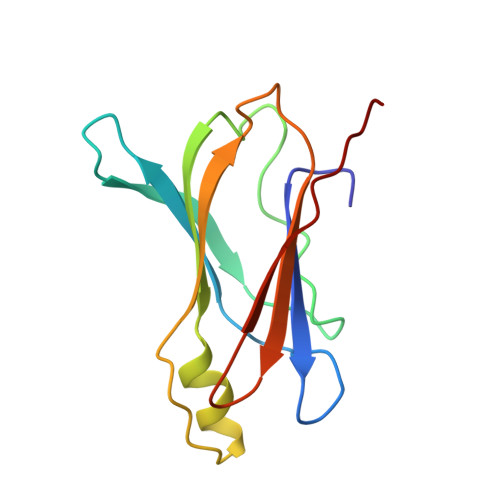
Crystal structure of rat transthyretin at 2.5 A resolution: first report on a unique tetrameric structure
Wojtczak, A.(1997) Acta Biochim Pol 44: 505-517
- PubMed: 9511961
- Primary Citation of Related Structures:
1GKE - PubMed Abstract:
The first observation of a unique tetrameric molecular structure of transthyretin from rat (rTTR, prealbumin) is reported. The structure has been determined by X-ray diffraction using molecular replacement and the structure of human transthyretin (hTTR) as a starting model. Crystals of native rat transthyretin are tetragonal, space group P4(3)2(1)2, and have four independent monomers in the asymmetric unit of the crystal lattice. Data were collected to 2.5 A resolution and the structure has been refined to R = 18.9% for 13584 data points between 8-2.5 A resolution. Like hTTR, the rat protein is also a 54000 Da tetramer with four identical polypeptide chains of 127 amino-acid residues. Of the 22 amino-acid residues which are different in the human and rat TTR sequences, none are in the thyroxine binding domain. Analysis of these data reveal that the tertiary structure of rTTR is similar to that of hTTR with only small differences in the flexible loop regions on the surface of the protein. As a result of local changes in flexible loop regions near residues 30-41, 60-65 and 102-104, the structure of rTTR monomers is more compact than that of the corresponding hTTR monomers. The loop between residues 30-41 is bound closer to the monomer core in the former as compared with the latter structure and there is a wider opening of the space formed between these loops at two adjacent monomeric subunits. These conformational changes do not affect the interfaces between the monomeric subunits and are not transmitted to the thyroxine binding site so that its topology remains not altered.
Organizational Affiliation:
Faculty of Chemistry, N. Copernicus University, Toruń, Poland.