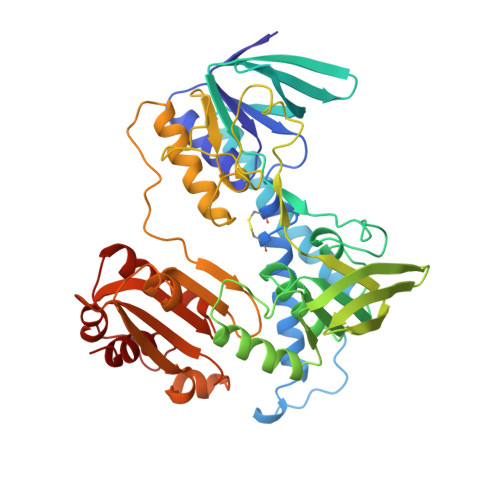
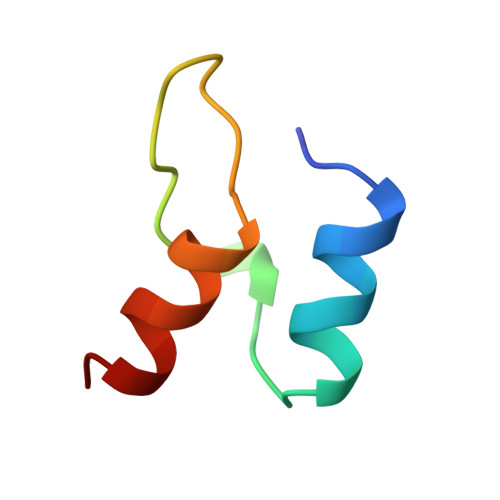
Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase.
Mande, S.S., Sarfaty, S., Allen, M.D., Perham, R.N., Hol, W.G.(1996) Structure 4: 277-286
- PubMed: 8805537
- DOI: https://doi.org/10.1016/s0969-2126(96)00032-9
- Primary Citation of Related Structures:
1EBD - PubMed Abstract:
The ubiquitous pyruvate dehydrogenase multienzyme complex is built around an octahedral or icosahedral core of dihydrolipoamide acetyltransferase (E2) chains, to which multiple copies of pyruvate decarboxylase (E1) and dihydrolipoamide dehydrogenase (E3) bind tightly but non-covalently. E2 is a flexible multidomain protein that mediates interactions with E1 and E3 through a remarkably small binding domain (E2BD).
Organizational Affiliation:
Department of Biological Structure, Biomolecular Structure Center, University of Washington, Box 357742, Seattle, WA 98195, USA.