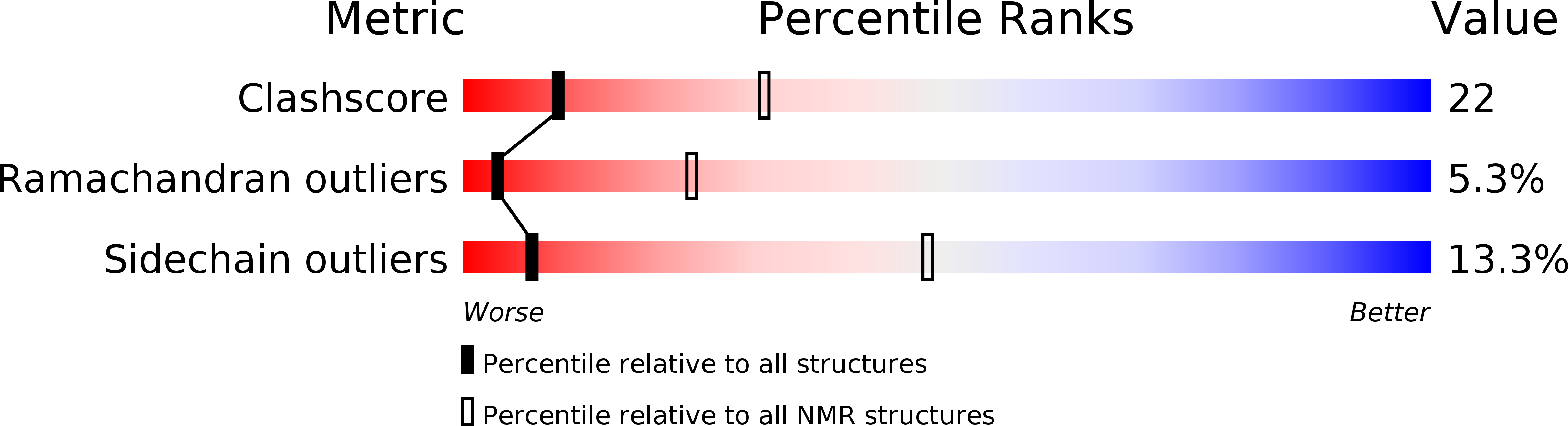Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
Morelli, X., Czjzek, M., Hatchikian, C.E., Bornet, O., Fontecilla-Camps, J.C., Palma, N.P., Moura, J.J., Guerlesquin, F.(2000) J Biol Chem 275: 23204
- PubMed: 10748163
- DOI: https://doi.org/10.1074/jbc.M909835199
- Primary Citation of Related Structures:
1E08 - PubMed Abstract:
Fe-hydrogenase is a 54-kDa iron-sulfur enzyme essential for hydrogen cycling in sulfate-reducing bacteria. The x-ray structure of Desulfovibrio desulfuricans Fe-hydrogenase has recently been solved, but structural information on the recognition of its redox partners is essential to understand the structure-function relationships of the enzyme. In the present work, we have obtained a structural model of the complex of Fe-hydrogenase with its redox partner, the cytochrome c(553), combining docking calculations and NMR experiments. The putative models of the complex demonstrate that the small subunit of the hydrogenase has an important role in the complex formation with the redox partner; 50% of the interacting site on the hydrogenase involves the small subunit. The closest contact between the redox centers is observed between Cys-38, a ligand of the distal cluster of the hydrogenase and Cys-10, a ligand of the heme in the cytochrome. The electron pathway from the distal cluster of the Fe-hydrogenase to the heme of cytochrome c(553) was investigated using the software Greenpath and indicates that the observed cysteine/cysteine contact has an essential role. The spatial arrangement of the residues on the interface of the complex is very similar to that already described in the ferredoxin-cytochrome c(553) complex, which therefore, is a very good model for the interacting domain of the Fe-hydrogenase-cytochrome c(553).
Organizational Affiliation:
Unité de Bioénergétique et Ingénierie des Protéines, IBSM-CNRS, Marseille Cedex 20, France.