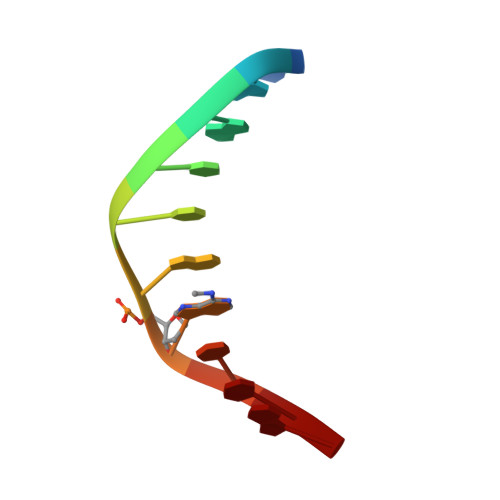
The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
Baikalov, I., Grzeskowiak, K., Yanagi, K., Quintana, J., Dickerson, R.E.(1993) J Mol Biol 231: 768-784
- PubMed: 8515450
- DOI: https://doi.org/10.1006/jmbi.1993.1325
- Primary Citation of Related Structures:
1DA3 - PubMed Abstract:
The B-DNA decanucleotide C-G-A-T-C-G-6meA-T-C-G has been crystallized under the same conditions used earlier for C-G-A-T-C-G-A-T-C-G, but is found to adopt a new trigonal P3(2)21 packing mode instead of the expected orthorhombic P2(1)2(1)2(1) form. Unit cell dimensions a = b = 33.38 A, c = 98.30 A, gamma = 120 degrees, imply ten base-pairs or one complete decamer double helix per asymmetric unit. The 2282 two-sigma data to 2.0 A refine to R = 17.2% with 45 water molecules, 1.5 hexavalent hydrated magnesium complexes, and 0.5 chloride ion per asymmetric unit. Neighboring helices interlock backbone chains and major grooves, crossing at an angle of 120 degrees in a manner that yields an excellent model for a Holliday junction. Local helix parameters differ markedly in the trigonal and orthorhombic structures, with the trigonal helix exhibiting behavior closer to that expected of B-DNA in solution. The trigonal form has an average of 10.6 base-pairs per turn, in contrast to 9.7 base-pairs per turn in the orthorhombic cell. A comparison of all known B-DNA decamer and dodecamer crystal structure analyses indicates that, the greater the cell volume per base-pair (and hence the more open the crystal structure), the closer the mean helix twist approaches an expected 10.6 base-pairs per turn.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Los Angeles 90024.