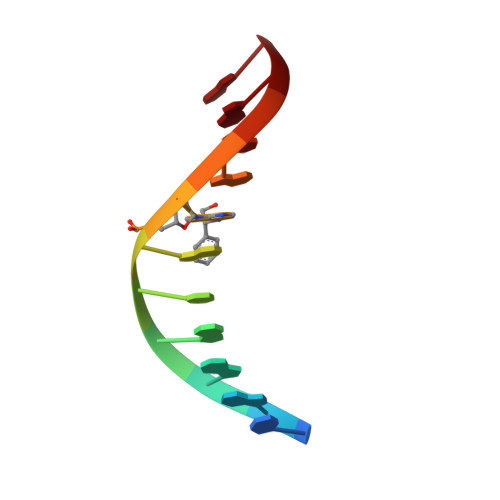
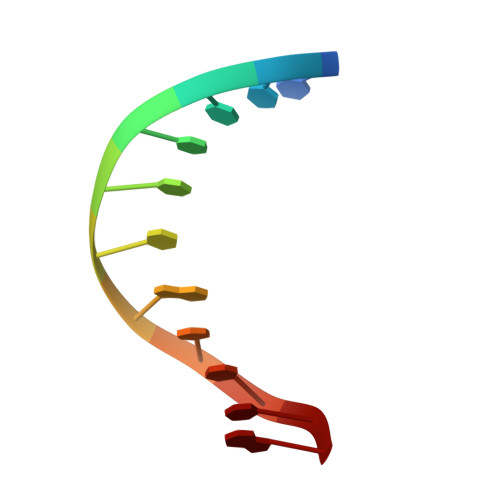
Major groove (R)-alpha-(N6-adenyl)styrene oxide adducts in an oligodeoxynucleotide containing the human N-ras codon 61 sequence: conformations of the R(61,2) and R(61,3) sequence isomers from 1H NMR.
Feng, B., Zhou, L., Passarelli, M., Harris, C.M., Harris, T.M., Stone, M.P.(1995) Biochemistry 34: 14021-14036
- PubMed: 7578000
- DOI: https://doi.org/10.1021/bi00043a008
- Primary Citation of Related Structures:
1AGK, 1AGZ - PubMed Abstract:
Conformations of (R)-alpha-(N6-adenyl)styrene oxide adducts at positions X6 in d(CGGACXAGAAG).d(CTTCTTGTCCG) and X7 in d(CGGACAXGAAG).d(CTTCTTGTCCG), incorporating codons 60, 61 (underlined), and 62 of the human n-ras protooncogene, were refined from 1H NMR data. These were the R(61,2) and R(61,3) adducts. Chemical shift perturbations were in the 5'-direction from the sites of adduction; large changes were observed for C5 H5 and H6 in the R(61,2) adduct. The styrene moieties were only partially defined by NOE data. Spectral overlap, particularly for the R(61,2) adduct, prevented complete assignments of the aromatic resonances; likewise, there were insufficient data to orient the CH2OH moieties. Ring flips were slow on the NMR time scale. For the R(61,2) adduct 260 restraints were obtained from NOE data at three mixing times using relaxation matrix analysis; for the R(61,3) adduct 230 restraints were obtained. Structures emergent from molecular dynamics/simulated annealing for the R(61,2) adduct converged to average and maximum pairwise rms differences of 1.3 and 1.7 A, respectively, while those for the R(61,3) adduct converged to average and maximum pairwise rms differences of 1.2 and 1.6 A. Sixth root residual indices of 7.5 and 6.8 x 10(-2) were measured between the refined structures and NOE intensities using relaxation matrix calculations for the R(61,2) and R(61,3) adducts, respectively. The styrene rings were in the 5'-direction from the lesion sites in the major groove. The preferred orientation calculated for the R(61,2) adduct placed the styrene ring edgewise and approximately orthogonal to C5, while that calculated for the R(61,3) duplex had the styrene ring approximately orthogonal to the major groove edges of base pairs A6.T17 and R-SOA7.T16.
Organizational Affiliation:
Center in Molecular Toxicology, Vanderbilt University, Nashville, Tennessee 37235, USA.