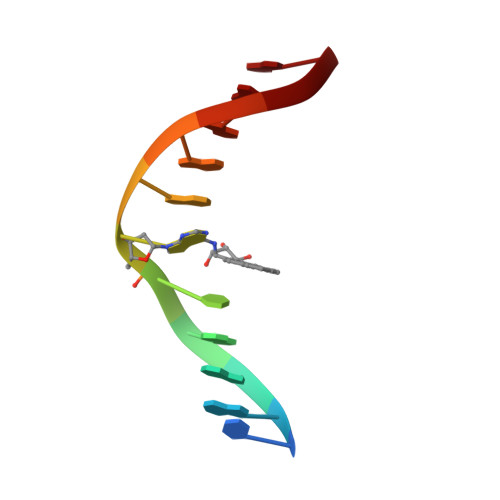
Adduction of the human N-ras codon 61 sequence with (-)-(7S,8R,9R,10S)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a] pyrene: structural refinement of the intercalated SRSR(61,2) (-)-(7S,8R,9S,1 0R)-N6-[10-(7,8,9,10- tetrahydrobenzo[a]pyrenyl)]-2'-deoxyadenosyl adduct from 1H NMR
Zegar, I.S., Kim, S.J., Johansen, T.N., Horton, P.J., Harris, C.M., Harris, T.M., Stone, M.P.(1996) Biochemistry 35: 6212-6224
- PubMed: 8639561
- DOI: https://doi.org/10.1021/bi9524732
- Primary Citation of Related Structures:
1AGU - PubMed Abstract:
The structure of the (-)-(7S,8R,9S,10R)-N6-[10-(7,8,910-tetrahydrobenzo [a]pyrenyl)]-2'-deoxyadenosyl adduct at X6 of 5'-d(CGGACXAGAAG)-3'-5'-d(CTTCTTGTCCG)-3', derived from trans addition of the exocyclic N6-amino group of dA to (-)-(7S,8R,9R,10S)-7, 8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene [(-)-DE2], was determined using molecular dynamics simulations restrained by 369 NOEs from 1H NMR. This was named the SRSR(61,2) adduct, derived from the N-ras protooncogene at and adjacent to the nucleotides encoding amino acid 61 (underlined) of the p21 gene product. NOEs between C5, S.R.S.R A6, and A7 were disrupted, as were those between T17 and G18. NOEs between benzo[a]pyrene and DNA protons were localized on the two faces of the pyrenyl ring. The benzo[a]pyrene H3-H6 protons showed NOEs to T17 CH3, while H1, H2, and H3 showed NOEs to T17 deoxyribose; the latter protons and H4 showed NOEs to T17 H2', H2" and to T17 H6. Noes were observed between H11 and H12 and C5 H]',H2', H2". G18 N1H showed NOEs to both faces of benzo[a]pyrene. Upfield shifts of 2.6 ppm for T17 N3H and 1.8 ppm for G18 N1H. 1 ppm for T17 H6 and CH3, and 0.75 ppm for C5 H5, with a smaller shift for C5 H6, and a 1.5 ppm dispersion of the pyrenyl protons suggested that benzo[a]pyrene intercalated above the 5'-face of S.R.S.R A6. The precision of the refined structures was monitored by pairwise root mean square deviations. which were < 1.5 A; accuracy was measured by complete relaxation matrix calculations, which yielded a sixth root R factor of 8.1 x 10(-2). Interstrand stacking between the pyrenyl ring and the T17 pyrimidine and G18 purine rings was enhanced by the bay ring. Changes of +30 degrees and -25 degrees in buckle for C5.G18 and S.R.S.R A6.T17, respectively, were calculated, as was a -40 degrees change in propeller twist for C5.G18. The rise between C5.G18 and S.R.S.R A6.T17 was calculated to be 7 A. The work extended the pattern for adenine N6 benzo[a]pyrene adducts, in which the R stereochemistry at C10 predicted 5'-intercalation of the pyrenyl moiety.
Organizational Affiliation:
Center in Molecular Toxicology, Vanderbilt University, Nashville, Tennessee 37235, USA.