Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Gallego, O., Ruiz, F.X., Ardevol, A., Dominguez, M., Alvarez, R., de Lera, A.R., Rovira, C., Farres, J., Fita, I., Pares, X.(2007) Proc Natl Acad Sci U S A 104: 20764-20769
- PubMed: 18087047
- DOI: https://doi.org/10.1073/pnas.0705659105
- Primary Citation of Related Structures:
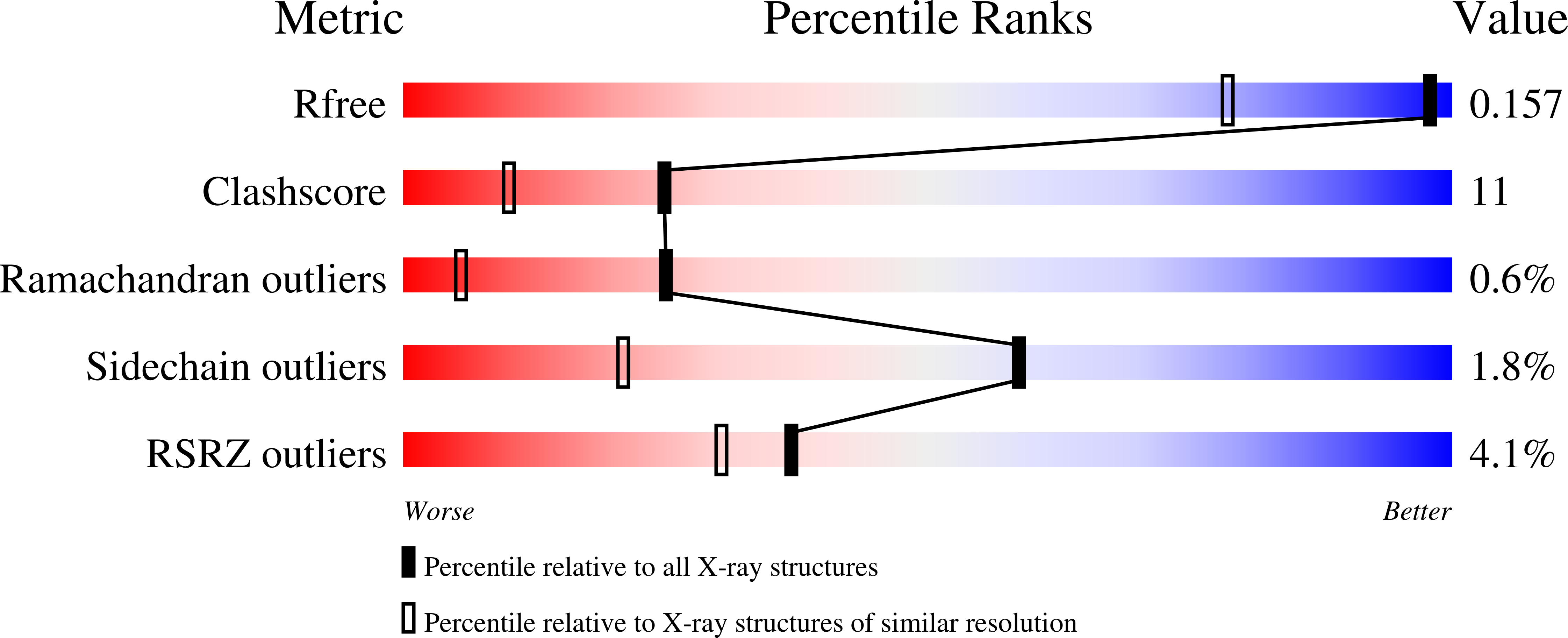
1ZUA - PubMed Abstract:
AKR1B10 is a human aldo-keto reductase (AKR) found to be elevated in several cancer types and in precancerous lesions. In vitro, AKR1B10 exhibits a much higher retinaldehyde reductase activity than any other human AKR, including AKR1B1 (aldose reductase). We here demonstrate that AKR1B10 also acts as a retinaldehyde reductase in vivo. This activity may be relevant in controlling the first step of retinoic acid synthesis. Up-regulation of AKR1B10, resulting in retinoic acid depletion, may lead to cellular proliferation. Both in vitro and in vivo activities of AKR1B10 were inhibited by tolrestat, an AKR1B1 inhibitor developed for diabetes treatment. The crystal structure of the ternary complex AKR1B10-NADP(+)-tolrestat was determined at 1.25-A resolution. Molecular dynamics models of AKR1B10 and AKR1B1 with retinaldehyde isomers and site-directed mutagenesis show that subtle differences at the entrance of the retinoid-binding site, especially at position 125, are determinant for the all-trans-retinaldehyde specificity of AKR1B10. Substitutions in the retinaldehyde cyclohexene ring also influence the specificity. These structural features should facilitate the design of specific inhibitors, with potential use in cancer and diabetes treatments.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Universitat Autònoma de Barcelona, E-08193 Bellaterra, Barcelona, Spain.