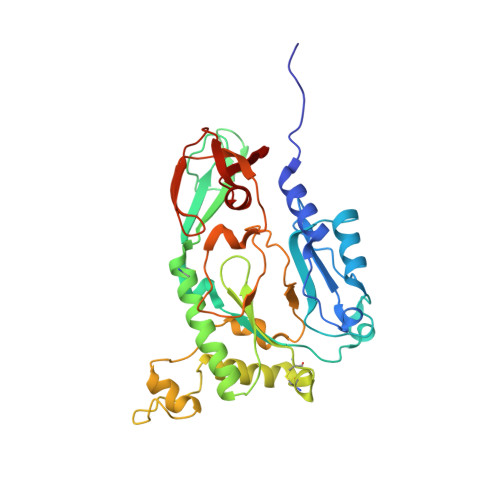
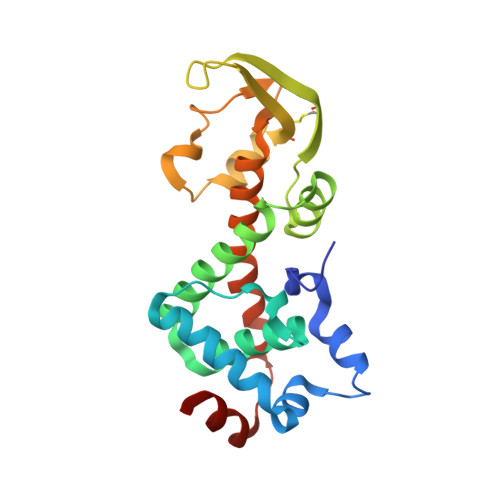
Structure of the heterodimeric core primase.
Lao-Sirieix, S.H., Nookala, R.K., Roversi, P., Bell, S.D., Pellegrini, L.(2005) Nat Struct Mol Biol 12: 1137-1144
- PubMed: 16273105
- DOI: https://doi.org/10.1038/nsmb1013
- Primary Citation of Related Structures:
1ZT2 - PubMed Abstract:
Primases are DNA-dependent RNA polymerases that synthesize the oligoribonucleotide primers essential to DNA replication. In archaeal and eukaryotic organisms, the core primase is a heterodimeric enzyme composed of a small and a large subunit. Here we report a crystallographic and biochemical analysis of the core primase from the archaeon Sulfolobus solfataricus. The structure provides the first three-dimensional description of the large subunit and its interaction with the small subunit. The evolutionary conservation of amino acids at the protein-protein interface implies that the observed mode of subunit association is conserved among archaeal and eukaryotic primases. The orientation of the large subunit in the core primase probably excludes its direct involvement in catalysis. Modeling of a DNA-RNA helix together with structure-based site-directed mutagenesis provides insight into the mechanism of template DNA binding and RNA primer synthesis.
Organizational Affiliation:
MRC Cancer Cell Unit, Hutchison MRC Research Centre, Hills Road, Cambridge CB2 2XZ, UK.