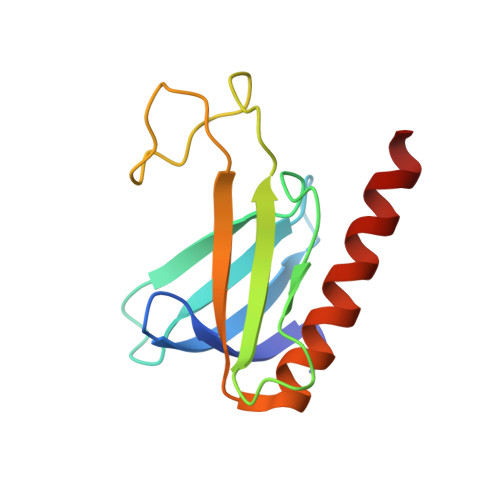
NMR Structure of the Amino-Terminal Domain from the Tfb1 Subunit of TFIIH and Characterization of Its Phosphoinositide and VP16 Binding Sites
Di Lello, P., Nguyen, B.D., Jones, T.N., Potempa, K., Kobor, M.S., Legault, P., Omichinski, J.G.(2005) Biochemistry 44: 7678-7686
- PubMed: 15909982
- DOI: https://doi.org/10.1021/bi050099s
- Primary Citation of Related Structures:
1Y5O - PubMed Abstract:
General transcription factor IIH (TFIIH) is recruited to the preinitiation complex (PIC) through direct interactions between its p62 (Tfb1) subunit and the carboxyl-terminal domain of TFIIEalpha. TFIIH has also been shown to interact with a number of transcriptional activator proteins through interactions with the same p62 (Tfb1) subunit. We have determined the NMR solution structure of the amino-terminal domain from the Tfb1 subunit of yeast TFIIH (Tfb1(1-115)). Like the corresponding domain from the human p62 protein, Tfb1(1-115) contains a PH domain fold despite a low level of sequence identity between the two functionally homologous proteins. In addition, we have performed in vitro binding studies that demonstrate that the PH domains of Tfb1 and p62 specifically bind to monophosphorylated inositides [PtdIns(5)P and PtdIns(3)P]. NMR chemical shift mapping demonstrated that the PtdIns(5)P binding site on Tfb1 (p62) is located in the basic pocket formed by beta-strands beta5-beta7 of the PH domain fold. Interestingly, the structural composition of the PtdIns(5)P binding site is different from the composition of the binding sites for phosphoinositides on prototypic PH domains. We have also determined that the PH domains from Tfb1 and p62 are sufficient for binding to the activation domain of VP16. NMR chemical shift mapping demonstrated that the VP16 binding site within the PH domain of Tfb1 (p62) overlaps with the PtdIns(5)P binding site on Tfb1 (p62). These results provide new information about the recognition of phosphoinositides by PH domains, and point to a potential role for phosphoinositides in VP16 regulation.
Organizational Affiliation:
Département de Biochimie, Université de Montréal, Montréal, Quebec, Canada.