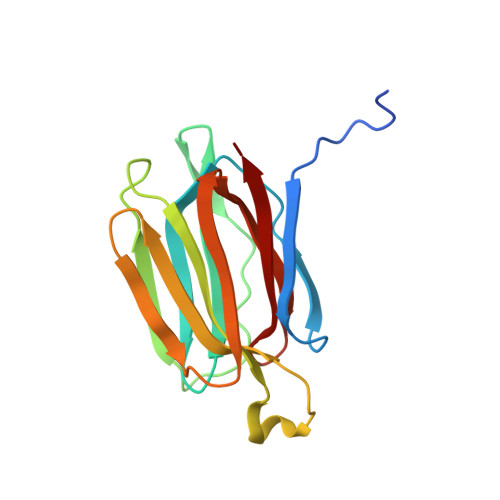
Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Rabijns, A., Barre, A., Van Damme, E.J.M., Peumans, W.J., De Ranter, C.J., Rouge, P.(2005) FEBS J 272: 3725-3732
- PubMed: 16008570
- DOI: https://doi.org/10.1111/j.1742-4658.2005.04801.x
- Primary Citation of Related Structures:
1XXQ, 1XXR - PubMed Abstract:
The structures of MornigaM and the MornigaM-mannose complex have been determined at 1.8 A and 2.0 A resolution, respectively. Both structures adopt the typical beta-prism motif found in other jacalin-related lectins and their tetrameric assembly closely resembles that of jacalin. The carbohydrate-binding cavity of MornigaM readily binds mannose. No major structural rearrangements can be observed in MornigaM upon binding of mannose. These results allow corroboration of the structure-function relationships within the small group of Moraceae lectins.
Organizational Affiliation:
Laboratory of Analytical Chemistry and Medicinal Physicochemistry, Faculty of Pharmaceutical Sciences, K. U. Leuven, Belgium.