Crystal structure of the histidine-containing phosphotransfer protein ZmHP2 from maize
Sugawara, H., Kawano, Y., Hatakeyama, T., Yamaya, T., Kamiya, N., Sakakibara, H.(2005) Protein Sci 14: 202-208
- PubMed: 15576555
- DOI: https://doi.org/10.1110/ps.041076905
- Primary Citation of Related Structures:
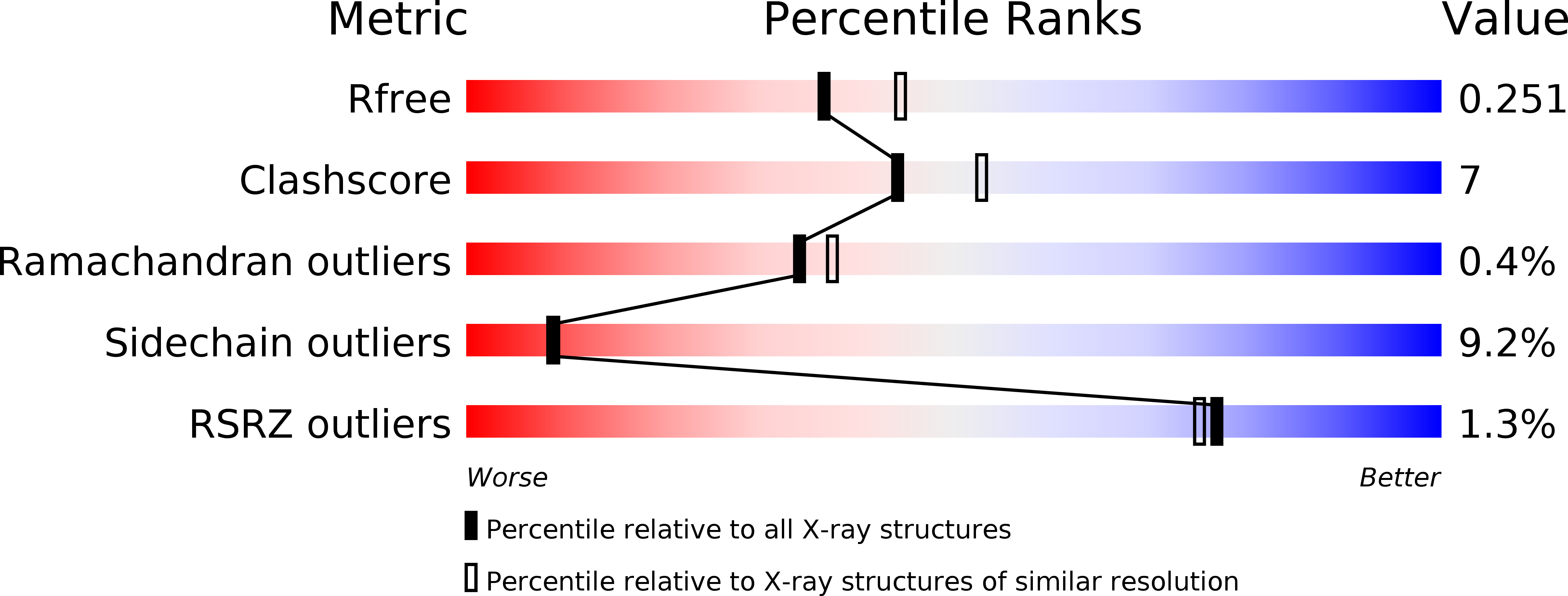
1WN0 - PubMed Abstract:
In higher plants, histidine-aspartate phosphorelays (two-component system) are involved in hormone signaling and stress responses. In these systems, histidine-containing phosphotransfer (HPt) proteins mediate the signal transmission from sensory histidine kinases to response regulators, including integration of several signaling pathways or branching into different pathways. We have determined the crystal structure of a maize HPt protein, ZmHP2, at 2.2 A resolution. ZmHP2 has six alpha-helices with a four-helix bundle at the C-terminus, a feature commonly found in HPt domains. In ZmHP2, almost all of the conserved residues among plant HPt proteins surround this histidine, probably forming the docking interface for the receiver domain of histidine kinase or the response regulator. Arg102 of ZmHP2 is conserved as a basic residue in plant HPt proteins. In bacteria, it is replaced by glutamine or glutamate that form a hydrogen bond to Ndelta atoms of the phospho-accepting histidine. It may play a key role in the complex formation of ZmHP2 with receiver domains.
Organizational Affiliation:
Laboratory for Communication Mechanisms, RIKEN Plant Science Center, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan. sugawara@psc.riken.go.jp