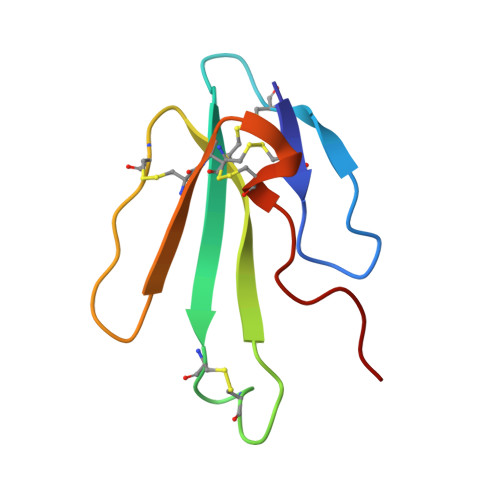
Solution structure of long neurotoxin NTX-1 from the venom of Naja naja oxiana by 2D-NMR spectroscopy.
Talebzadeh-Farooji, M., Amininasab, M., Elmi, M.M., Naderi-Manesh, H., Sarbolouki, M.N.(2004) Eur J Biochem 271: 4950-4957
- PubMed: 15606783
- DOI: https://doi.org/10.1111/j.1432-1033.2004.04465.x
- Primary Citation of Related Structures:
1W6B - PubMed Abstract:
The NMR solution structures of NTX-1 (PDB code 1W6B and BMRB 6288), a long neurotoxin isolated from the venom of Naja naja oxiana, and the molecular dynamics simulation of these structures are reported. Calculations are based on 1114 NOEs, 19 hydrogen bonds, 19 dihedral angle restraints and secondary chemical shifts derived from 1H to 13C HSQC spectrum. Similar to other long neurotoxins, the three-finger like structure shows a double and a triple stranded beta-sheet as well as some flexible regions, particularly at the tip of loop II and the C-terminal tail. The solution NMR and molecular dynamics simulated structures are in good agreement with root mean square deviation values of 0.23 and 1 A for residues involved in beta-sheet regions, respectively. The overall fold in the NMR structure is similar to that of the X-ray crystallography, although some differences exist in loop I and the tip of loop II. The most functionally important residues are located at the tip of loop II and it appears that the mobility and the local structure in this region modulate the binding of NTX-1 and other long neurotoxins to the nicotinic acetylcholine receptor.
Organizational Affiliation:
Institute of Biochemistry & Biophysics, University of Tehran, Iran.