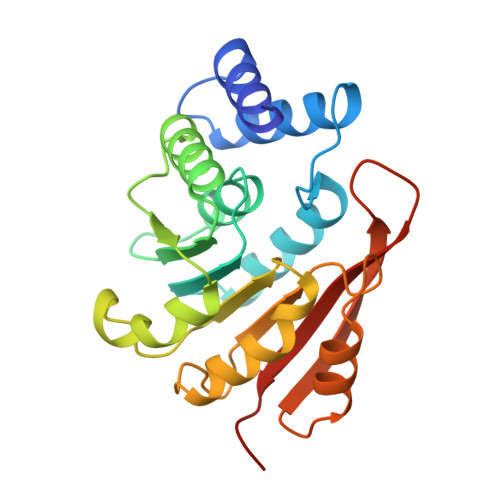
Crystal structure of catechol O-methyltransferase.
Vidgren, J., Svensson, L.A., Liljas, A.(1994) Nature 368: 354-358
- PubMed: 8127373
- DOI: https://doi.org/10.1038/368354a0
- Primary Citation of Related Structures:
1VID - PubMed Abstract:
Catechol O-methyltransferase (COMT, EC 2.1.1.6) is important in the central nervous system because it metabolizes catecholamine neurotransmitters such as dopamine. The enzyme catalyses the transfer of the methyl group from S-adenosyl-L-methionine (AdoMet) to one hydroxyl group of catechols. COMT also inactivates catechol-type compounds such as L-DOPA. With selective inhibitors of COMT in combination with L-DOPA, a new principle has been realized in the therapy of Parkinson's disease. Here we solve the atomic structure of COMT to 2.0 A resolution, which provides new insights into the mechanism of the methyl transfer reaction. The co-enzyme-binding domain is strikingly similar to that of an AdoMet-dependent DNA methylase, indicating that all AdoMet methylases may have a common structure.
Organizational Affiliation:
Orion Corporation, Espoo, Finland.