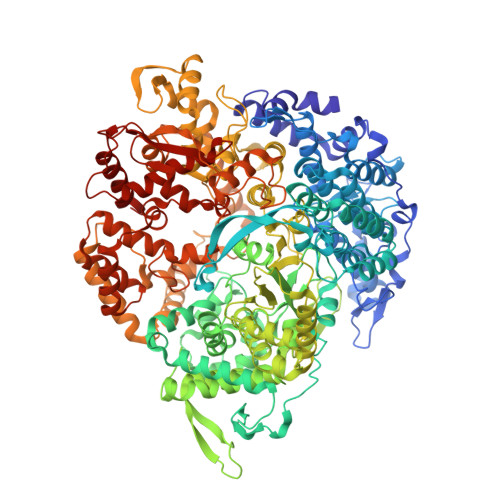
Reovirus Polymerase Lambda3 Localized by Cryo-Electron Microscopy of Virions at a Resolution of 7.6 A
Zhang, X., Walker, S.B., Chipman, P.R., Nibert, M.L., Baker, T.S.(2003) Nat Struct Biol 10: 1011
- PubMed: 14608373
- DOI: https://doi.org/10.1038/nsb1009
- Primary Citation of Related Structures:
1UON - PubMed Abstract:
Reovirus is an icosahedral, double-stranded (ds) RNA virus that uses viral polymerases packaged within the viral core to transcribe its ten distinct plus-strand RNAs. To localize these polymerases, the structure of the reovirion was refined to a resolution of 7.6 A by cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction. X-ray crystal models of reovirus proteins, including polymerase lambda 3, were then fitted into the density map. Each copy of lambda 3 was found anchored to the inner surface of the icosahedral core shell, making major contacts with three molecules of shell protein lambda 1 and overlapping, but not centering on, a five-fold axis. The overlap explains why only one copy of lambda 3 is bound per vertex. lambda 3 is furthermore oriented with its transcript exit channel facing a small channel through the lambda 1 shell, suggesting how the nascent RNA is passed into the large external cavity of the pentameric capping enzyme complex formed by protein lambda 2.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907, USA.