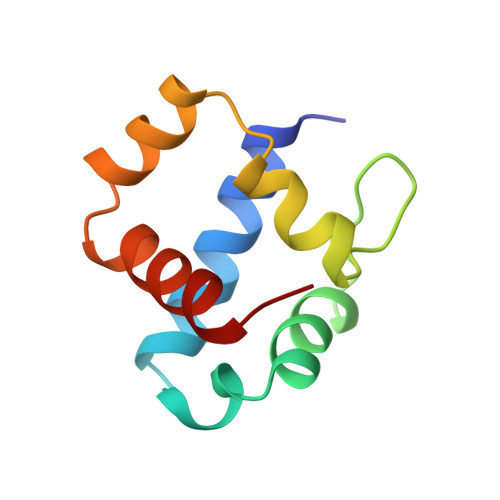
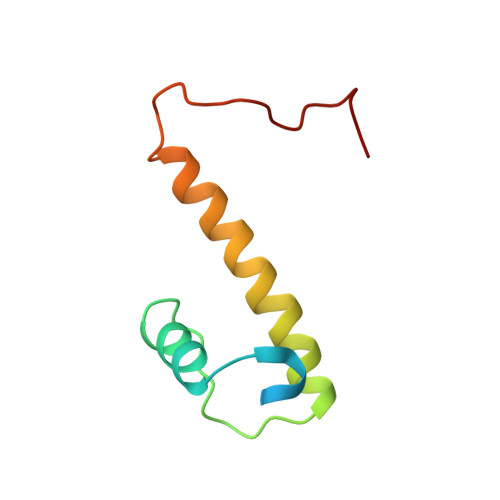
T4 AsiA blocks DNA recognition by remodeling sigma(70) region 4
Lambert, L.J., Wei, Y., Schirf, V., Demeler, B., Werner, M.H.(2004) EMBO J 23: 2952-2962
- PubMed: 15257291
- DOI: https://doi.org/10.1038/sj.emboj.7600312
- Primary Citation of Related Structures:
1TL6, 1TLH, 1TTY - PubMed Abstract:
Bacteriophage T4 AsiA is a versatile transcription factor capable of inhibiting host gene expression as an 'anti-sigma' factor while simultaneously promoting gene-specific expression of T4 middle genes in conjunction with T4 MotA. To accomplish this task, AsiA engages conserved region 4 of Eschericia coli sigma70, blocking recognition of most host promoters by sequestering the DNA-binding surface at the AsiA/sigma70 interface. The three-dimensional structure of an AsiA/region 4 complex reveals that the C-terminal alpha helix of region 4 is unstructured, while four other helices adopt a completely different conformation relative to the canonical structure of unbound region 4. That AsiA induces, rather than merely stabilizes, this rearrangement can be realized by comparison to the homologous structures of region 4 solved in a variety of contexts, including the structure of Thermotoga maritima sigmaA region 4 described herein. AsiA simultaneously occupies the surface of region 4 that ordinarily contacts core RNA polymerase (RNAP), suggesting that an AsiA-bound sigma70 may also undergo conformational changes in the context of the RNAP holoenzyme.
Organizational Affiliation:
Laboratory of Molecular Biophysics, Rockefeller University, New York, NY 10021, USA.