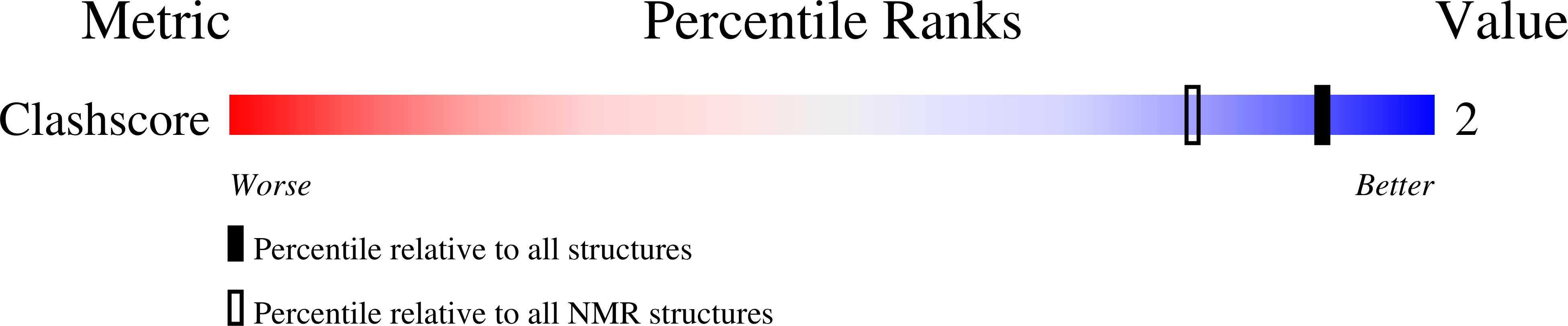Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
McAteer, K., Aceves-Gaona, A., Michalczyk, R., Buchko, G.W., Isern, N.G., Silks, L.A., Miller, J.H., Kennedy, M.A.(2004) Biopolymers 75: 497-511
- PubMed: 15526287
- DOI: https://doi.org/10.1002/bip.20168
- Primary Citation of Related Structures:
1SS7, 1SSV - PubMed Abstract:
In-phase ligated DNA containing T(n)A(n) segments fail to exhibit the retarded polyacrylamide gel electrophoresis (PAGE) migration observed for in-phase ligated A(n)T(n) segments, a behavior thought to be correlated with macroscopic DNA curvature. The lack of macroscopic curvature in ligated T(n)A(n) segments is thought to be due to cancellation of bending in regions flanking the TpA steps. To address this issue, solution-state NMR, including residual dipolar coupling (RDC) restraints, was used to determine a high-resolution structure of [d(CGAGGTTTAAACCTCG)2], a DNA oligomer containing a T3A3 tract. The overall magnitude and direction of bending, including the regions flanking the central TpA step, was measured using a radius of curvature, Rc, analysis. The Rc for the overall molecule indicated a small magnitude of global bending (Rc = 138 +/- 23 nm) towards the major groove, whereas the Rc for the two halves (72 +/- 33 nm and 69 +/- 14 nm) indicated greater localized bending into the minor groove. The direction of bending in the regions flanking the TpA step is in partial opposition (109 degrees), contributing to cancellation of bending. The cancellation of bending did not correlate with a pattern of roll values at the TpA step, or at the 5' and 3' junctions, of the T3A3 segment, suggesting a simple junction/roll model is insufficient to predict cancellation of DNA bending in all T(n)A(n) junction sequence contexts. Importantly, Rc analysis of structures refined without RDC restraints lacked the precision and accuracy needed to reliably measure bending.
Organizational Affiliation:
Department of Computer Science and Electrical Engineering, Washington State University Tri-Cities, Richland, WA 99352.