Alterations at the Peptidyl Transferase Centre of the Ribosome Induced by the Synergistic Action of the Streptogramins Dalfopristin and Quinupristin.
Harms, J.M., Schlunzen, F., Fucini, P., Bartels, H., Yonath, A.(2004) BMC Biol 2: 4
- PubMed: 15059283
- DOI: https://doi.org/10.1186/1741-7007-2-4
- Primary Citation of Related Structures:
1SM1 - PubMed Abstract:
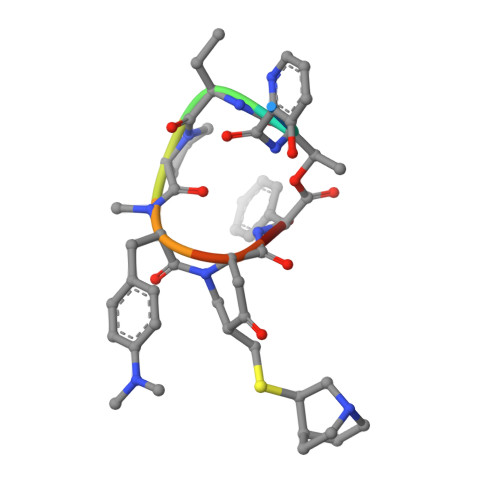
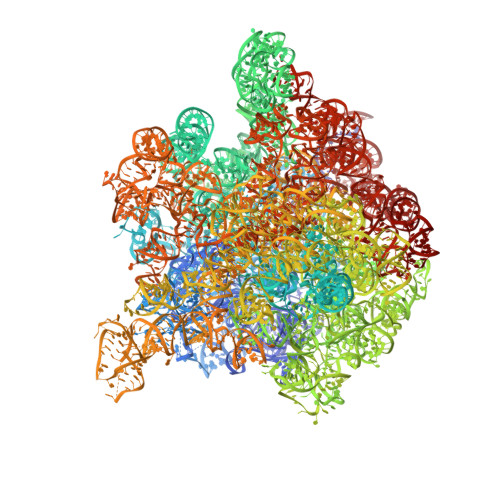
The bacterial ribosome is a primary target of several classes of antibiotics. Investigation of the structure of the ribosomal subunits in complex with different antibiotics can reveal the mode of inhibition of ribosomal protein synthesis. Analysis of the interactions between antibiotics and the ribosome permits investigation of the specific effect of modifications leading to antimicrobial resistances. Streptogramins are unique among the ribosome-targeting antibiotics because they consist of two components, streptogramins A and B, which act synergistically. Each compound alone exhibits a weak bacteriostatic activity, whereas the combination can act bactericidal. The streptogramins A display a prolonged activity that even persists after removal of the drug. However, the mode of activity of the streptogramins has not yet been fully elucidated, despite a plethora of biochemical and structural data. The investigation of the crystal structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with the clinically relevant streptogramins quinupristin and dalfopristin reveals their unique inhibitory mechanism. Quinupristin, a streptogramin B compound, binds in the ribosomal exit tunnel in a similar manner and position as the macrolides, suggesting a similar inhibitory mechanism, namely blockage of the ribosomal tunnel. Dalfopristin, the corresponding streptogramin A compound, binds close to quinupristin directly within the peptidyl transferase centre affecting both A- and P-site occupation by tRNA molecules. The crystal structure indicates that the synergistic effect derives from direct interaction between both compounds and shared contacts with a single nucleotide, A2062. Upon binding of the streptogramins, the peptidyl transferase centre undergoes a significant conformational transition, which leads to a stable, non-productive orientation of the universally conserved U2585. Mutations of this rRNA base are known to yield dominant lethal phenotypes. It seems, therefore, plausible to conclude that the conformational change within the peptidyl transferase centre is mainly responsible for the bactericidal activity of the streptogramins and the post-antibiotic inhibition of protein synthesis.
Organizational Affiliation:
Max-Planck Research Unit for Ribosomal Structure, 22603 Hamburg, Germany. harms@riboworld.com