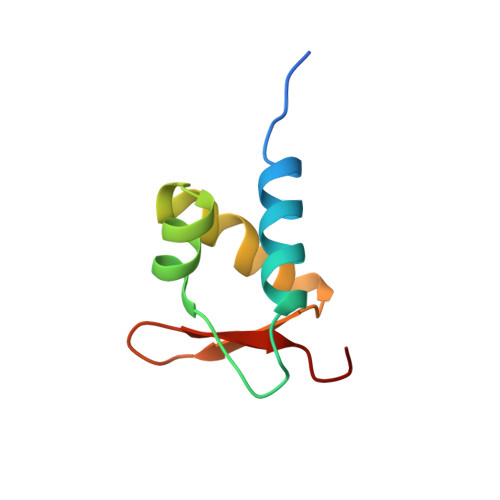
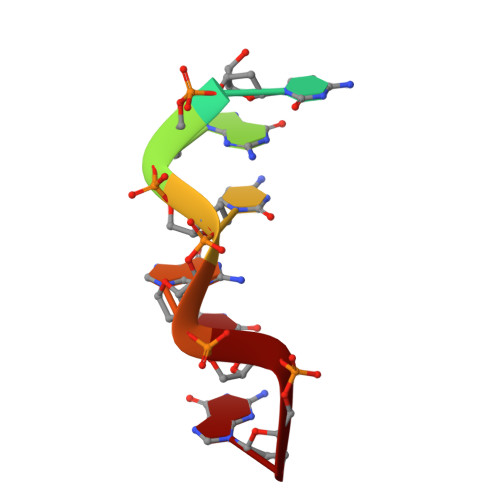
A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA.
Ha, S.C., Lokanath, N.K., Van Quyen, D., Wu, C.A., Lowenhaupt, K., Rich, A., Kim, Y.G., Kim, K.K.(2004) Proc Natl Acad Sci U S A 101: 14367-14372
- PubMed: 15448208
- DOI: https://doi.org/10.1073/pnas.0405586101
- Primary Citation of Related Structures:
1SFU - PubMed Abstract:
A conserved feature of poxviruses is a protein, well characterized as E3L in vaccinia virus, that confers IFN resistance on the virus. This protein comprises two domains, an N-terminal Z-DNA-binding protein domain (Zalpha) and a C-terminal double-stranded RNA-binding domain. Both are required for pathogenicity of vaccinia virus in mice infected by intracranial injection. Here, we describe the crystal structure of the Zalpha domain from the E3L-like protein of Yaba-like disease virus, a Yatapoxvirus, in a complex with Z-DNA, solved at a 2.0-A resolution. The DNA contacting surface of Yaba-like disease virus Zalpha(E3L) closely resembles that of other structurally defined members of the Zalpha family, although some variability exists in the beta-hairpin region. In contrast to the Z-DNA-contacting surface, the nonbinding surface of members of the Zalpha family are unrelated; this surface may effect protein-specific interactions. The presence of the conserved and tailored Z-DNA-binding surface, which interacts specifically with the zigzag backbone and syn base diagnostic of the Z-form, reinforces the importance to poxvirus infection of the ability of this protein to recognize the Z-conformation.
Organizational Affiliation:
Department of Molecular Cell Biology, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Suwon 440-746, Korea.