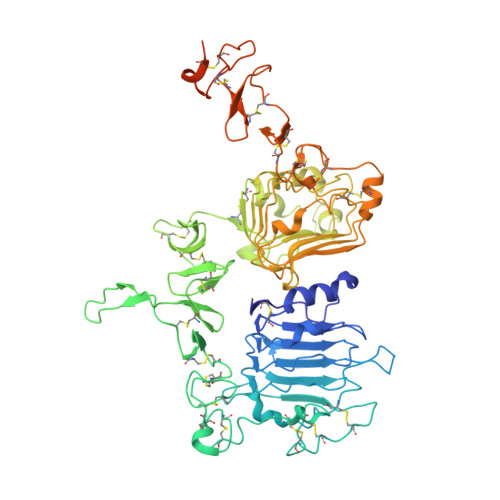
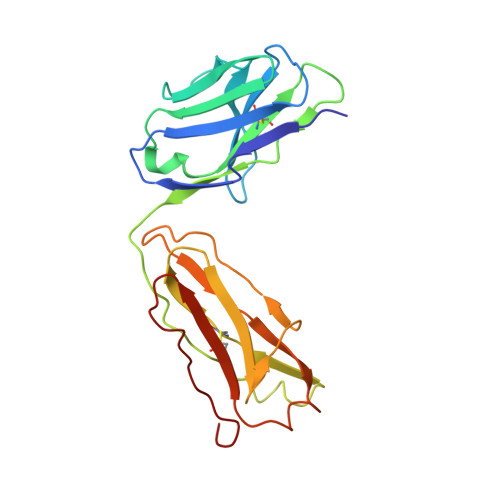
Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex.
Franklin, M.C., Carey, K.D., Vajdos, F.F., Leahy, D.J., De Vos, A.M., Sliwkowski, M.X.(2004) Cancer Cell 5: 317-328
- PubMed: 15093539
- DOI: https://doi.org/10.1016/s1535-6108(04)00083-2
- Primary Citation of Related Structures:
1S78 - PubMed Abstract:
We have determined the 3.2 A X-ray crystal structure of the extracellular domain of the human epidermal growth factor receptor 2 (ErbB2 or HER2) in a complex with the antigen binding fragment of pertuzumab, an anti-ErbB2 monoclonal antibody also known as 2C4 or Omnitarg. Pertuzumab binds to ErbB2 near the center of domain II, sterically blocking a binding pocket necessary for receptor dimerization and signaling. The ErbB2-pertuzumab structure, combined with earlier mutagenesis data, defines the pertuzumab residues essential for ErbB2 interaction. To analyze the ErbB2 side of the interface, we have mutated a number of residues contacting pertuzumab and examined the effects of these mutations on pertuzumab binding and ErbB2-ErbB3 heterodimerization. We have also shown that conserved residues previously shown to be necessary for EGF receptor homodimerization may be dispensible for ErbB2-ErbB3 heterodimerization.
Organizational Affiliation:
Department of Protein Engineering, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94114 USA.