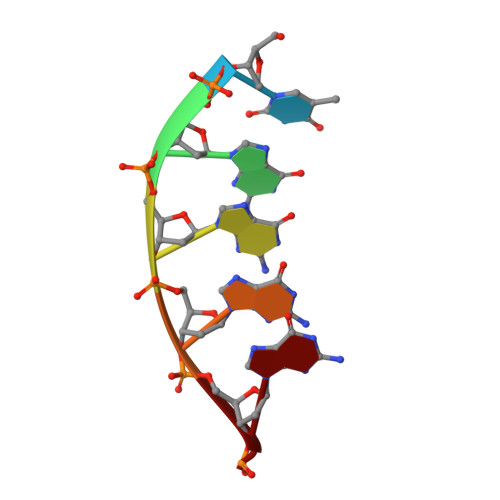
A Thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+1/Na+1 ions
Caceres, C., Wright, G., Gouyette, C., Parkinson, G., Subirana, J.A.(2004) Nucleic Acids Res 32: 1097-1102
- PubMed: 14960719
- DOI: https://doi.org/10.1093/nar/gkh269
- Primary Citation of Related Structures:
1S45, 1S47 - PubMed Abstract:
We report two new structures of the quadruplex d(TGGGGT)4 obtained by single crystal X-ray diffraction. In one of them a thymine tetrad is found. Thus the yeast telomere sequences d(TG1-3) might be able to form continuous quadruplex structures, involving both guanine and thymine tetrads. Our study also shows substantial differences in the arrangement of thymines when compared with previous studies. We find five different types of organization: (i) groove binding with hydrogen bonds to guanines from a neighbour quadruplex; (ii) partially ordered groove binding, without any hydrogen bond; (iii) stacked thymine triads, formed at the 3'ends of the quadruplexes; (iv) a thymine tetrad between two guanine tetrads. Thymines are stabilized in pairs by single hydrogen bonds. A central sodium ion interacts with two thymines and contributes to the tetrad structure. (v) Completely disordered thymines which do not show any clear location in the crystal. The tetrads are stabilized by either Na+ or Tl+ ions. We show that by using MAD methods, Tl+ can be unambiguously located and distinguished from Na+. We can thus determine the preference for either ion in each ionic site of the structure under the conditions used by us.
Organizational Affiliation:
Departament d'Enginyeria Química, Universitat Politècnica de Catalunya, Avinguda Diagonal 647, E-08028 Barcelona, Spain.