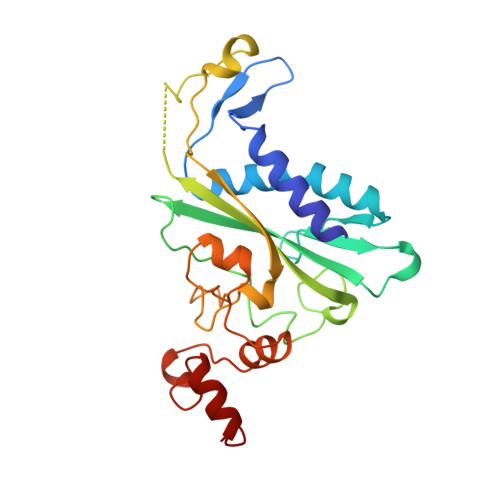
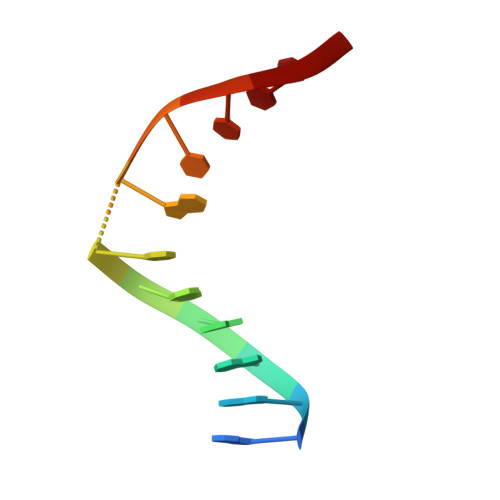
Role of protein-induced bending in the specificity of DNA recognition: crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + d(ATCTT).
Horton, N.C., Perona, J.J.(1998) J Mol Biol 277: 779-783
- PubMed: 9545372
- DOI: https://doi.org/10.1006/jmbi.1998.1655
- Primary Citation of Related Structures:
1RV5 - PubMed Abstract:
The crystal structure of EcoRV endonuclease has been determined at 2. 1 A resolution complexed to two five-base-pair DNA duplexes each containing the cognate recognition half-site. The highly localized 50 degrees bend into the major groove seen at the center TA-step of the continuous GATATC site is preserved in this discontinuous DNA complex lacking the scissile phosphates. Thus, this crystal structure provides evidence that covalent constraints associated with a continuous target site are not essential to enzyme-induced DNA bending, even when these constraints are removed directly at the locus of the bend. The scissile phosphates are also absent in the crystal structure of EcoRV bound to the non-specific site TCGCGA, which shows a straight B-like conformation. We conclude that DNA bending by EcoRV is governed only by the sequence and is not influenced by the continuity of the phosphodiester backbone. Together with other data showing that cleavable non-cognate sites are bent, these results indicate that EcoRV bends non-cognate sites differing by one or two base-pairs from GATATC, but does not bend non-specific sites that are less similar. Structural and thermodynamic considerations suggest that the sequence-dependent energy cost of DNA bending is likely to play an important role in determining the specificity of EcoRV. This differential cost is manifested at the binding step for bent non-cognate sequences and at the catalytic step for unbent non-specific sequences.
Organizational Affiliation:
Department of Chemistry and Interdepartmental Program in Biochemistry and Molecular Biology, University of California at Santa Barbara, Santa Barbara, CA, 93106-9510, USA.