Three-dimensional structure of an unusual Kunitz (STI) type trypsin inhibitor from Copaifera langsdorffii.
Krauchenco, S., Nagem, R.A.P., Da Silva, J.A., Marangoni, S., Polikarpov, I.(2004) Biochimie 86: 167-172
- PubMed: 15134830
- DOI: https://doi.org/10.1016/j.biochi.2004.03.004
- Primary Citation of Related Structures:
1R8O - PubMed Abstract:
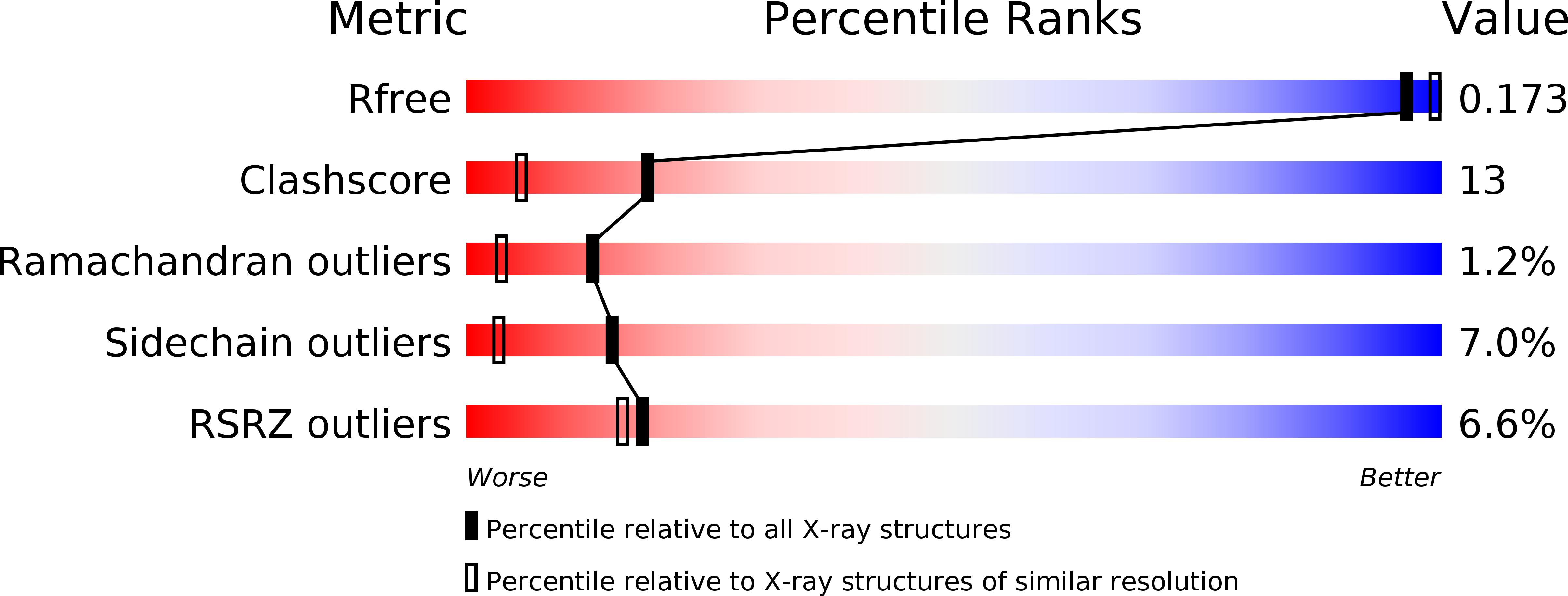
The crystallographic structure of a novel trypsin inhibitor (CTI) from Copaifera langsdorffii is reported. The structure was solved by MIRAS procedure and refined to a crystallographic residual of 17.3% (R(free) = 20.3%) at 1.8 A resolution. Two isomorphous derivatives were obtained by quick cryo-soaking approach. CTI is the first structure of a member of Kunitz (STI) family formed by two noncovalently bound polypeptide chains and only one disulfide bridge. A standard Kunitz-type inhibitor has a single polypeptide chain and two disulfide bridges. Structural features granting CTI high inhibitory activity are discussed.
Organizational Affiliation:
Grupo de Cristalografia, Department of Physics and Informatics, Institute of Physics (IFSC), University of Sao Paulo, Avenue Trabalhador Saocarlense, 400, CEP 13560-970, Sao Carlos, SP, Brazil.