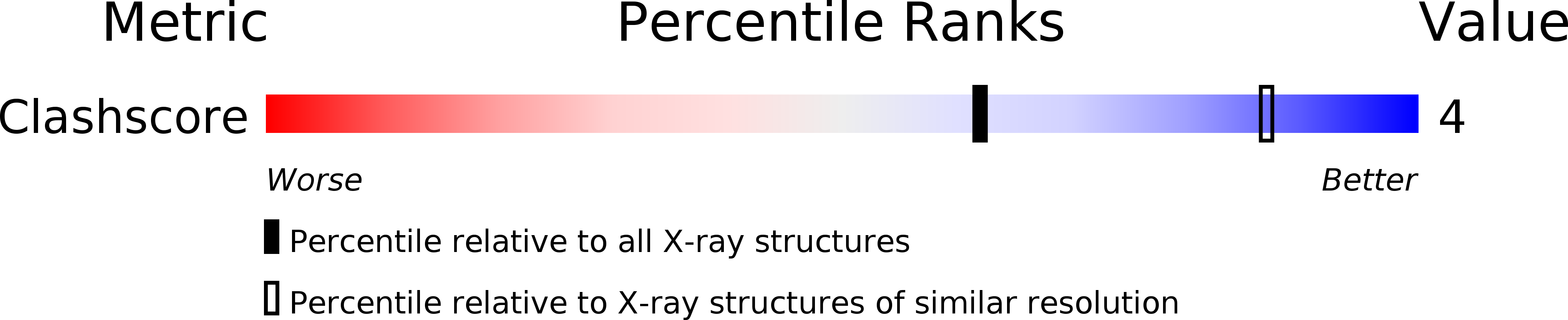Solution Structure Determination of Monomeric Human IgA2 by X-ray and Neutron Scattering, Analytical Ultracentrifugation and Constrained Modelling: A Comparison with Monomeric Human IgA1.
Furtado, P.B., Whitty, P.W., Robertson, A., Eaton, J.T., Almogren, A., Kerr, M.A., Woof, J.M., Perkins, S.J.(2004) J Mol Biol 338: 921-941
- PubMed: 15111057
- DOI: https://doi.org/10.1016/j.jmb.2004.03.007
- Primary Citation of Related Structures:
1R70 - PubMed Abstract:
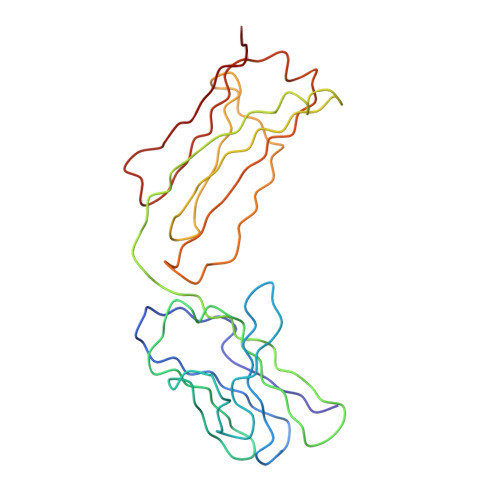
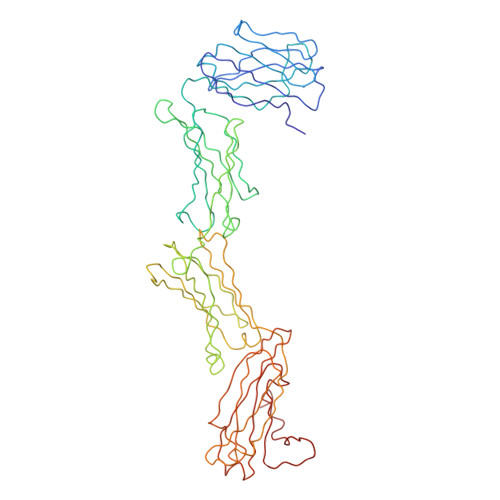
Immunoglobulin A (IgA), the most abundant human immunoglobulin, mediates immune protection at mucosal surfaces as well as in plasma. It exists as two subclasses IgA1 and IgA2, and IgA2 is found in at least two allotypic forms, IgA2m(1) or IgA2m(2). Compared to IgA1, IgA2 has a much shorter hinge region, which joins the two Fab and one Fc fragments. In order to assess its solution structure, monomeric recombinant IgA2m(1) was studied by X-ray and neutron scattering. Its Guinier X-ray radius of gyration R(G) is 5.18 nm and its neutron R(G) is 5.03 nm, both of which are significantly smaller than those for monomeric IgA1 at 6.1-6.2 nm. The distance distribution function P(r)for IgA2m(1) showed a broad peak with a subpeak and gave a maximum dimension of 17 nm, in contrast to the P(r) curve for IgA1, which showed two distinct peaks and a maximum dimension of 21 nm. The sedimentation coefficients of IgA1 and IgA2m(1) were 6.2S and 6.4S, respectively. These data show that the solution structure of IgA2m(1) is significantly more compact than IgA1. The complete monomeric IgA2m(1) structure was modelled using molecular dynamics to generate random IgA2 hinge structures, to which homology models for the Fab and Fc fragments were connected to generate 10,000 full models. A total of 104 compact best-fit IgA2m(1) models gave good curve fits. These best-fit models were modified by linking the two Fab light chains with a disulphide bridge that is found in IgA2m(1), and subjecting these to energy refinement to optimise this linkage. The averaged solution structure of the arrangement of the Fab and Fc fragments in IgA2m(1) was found to be predominantly T-shaped and flexible, but also included Y-shaped structures. The IgA2 models show full steric access to the two FcalphaRI-binding sites at the Calpha2-Calpha3 interdomain region in the Fc fragment. Since previous scattering modelling had shown that IgA1 also possessed a flexible T-shaped solution structure, such a T-shape may be common to both IgA1 and IgA2. The final models suggest that the combination of the more compact IgA2m(1) and the more extended IgA1 structures will enable human IgA to access a broader range of antigens than either acting alone. The hinges of both IgA subclasses appear to show reduced flexibility when compared to their equivalents in IgG, and this may be important for maintaining an extended IgA structure.
Organizational Affiliation:
Structural Immunology Group, Department of Biochemistry and Molecular Biology, Royal Free and University College Medical School, University College London, Gower Street, London WC1E 6BT, UK.