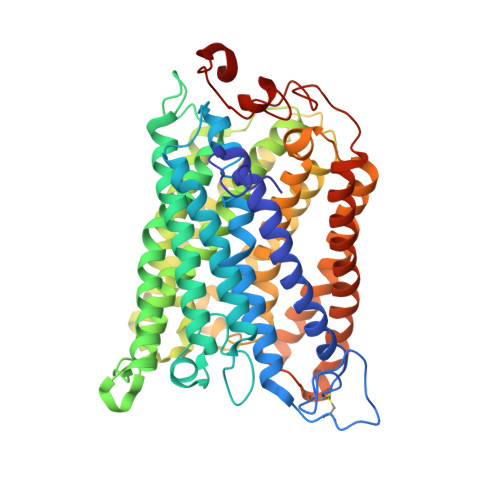
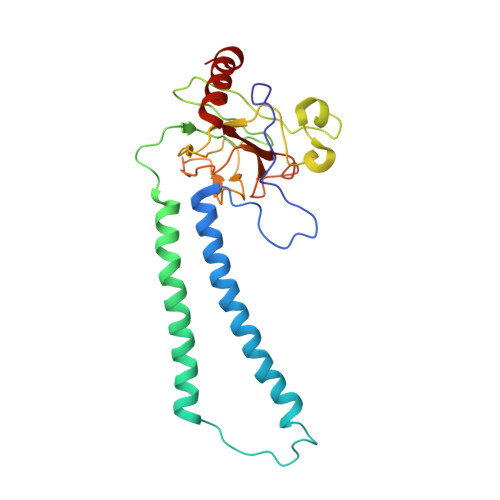
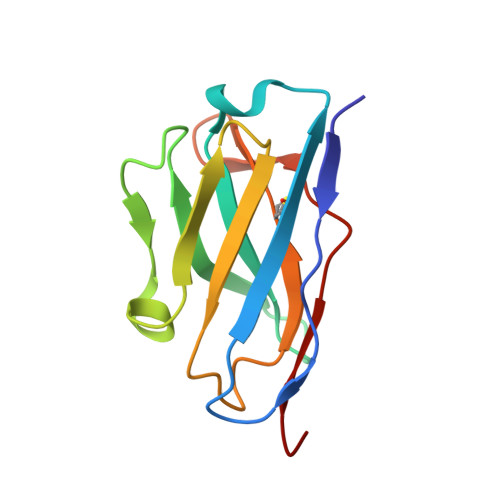
The Cytochrome C Oxidase from Paracoccus Denitrificans Does not Change the Metal Center Ligation Upon Reduction
Harrenga, A., Michel, H.(1999) J Biol Chem 274: 33296
- PubMed: 10559205
- DOI: https://doi.org/10.1074/jbc.274.47.33296
- Primary Citation of Related Structures:
1QLE - PubMed Abstract:
Cytochrome c oxidase catalyzes the reduction of oxygen to water. This process is accompanied by the vectorial transport of protons across the mitochondrial or bacterial membrane ("proton pumping"). The mechanism of proton pumping is still a matter of debate. Many proposed mechanisms require structural changes during the reaction cycle of cytochrome c oxidase. Therefore, the structure of the cytochrome c oxidase was determined in the completely oxidized and in the completely reduced states at a temperature of 100 K. No ligand exchanges or other major structural changes upon reduction of the cytochrome c oxidase from Paracoccus denitrificans were observed. The three histidine Cu(B) ligands are well defined in the oxidized and in the reduced states. These results are hardly compatible with the "histidine cycle" mechanisms formulated previously.
Organizational Affiliation:
Max-Planck-Institut für Biophysik, Abteilung für molekulare Membranbiologie, Heinrich-Hoffmann-Strasse 7, 60528 Frankfurt am Main, Germany.