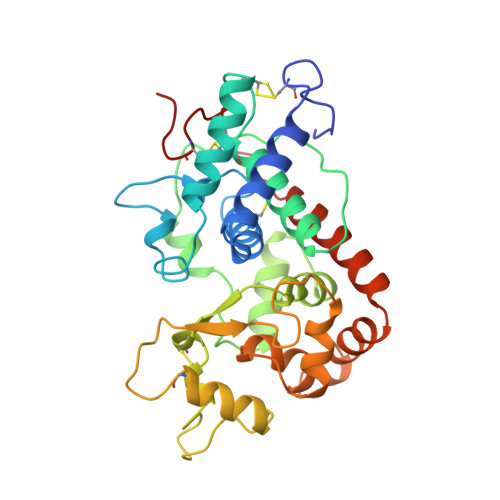
Arabidopsis thaliana peroxidase N: structure of a novel neutral peroxidase.
Mirza, O., Henriksen, A., Ostergaard, L., Welinder, K.G., Gajhede, M.(2000) Acta Crystallogr D Biol Crystallogr 56: 372-375
- PubMed: 10713531
- DOI: https://doi.org/10.1107/s0907444999016340
- Primary Citation of Related Structures:
1QGJ - PubMed Abstract:
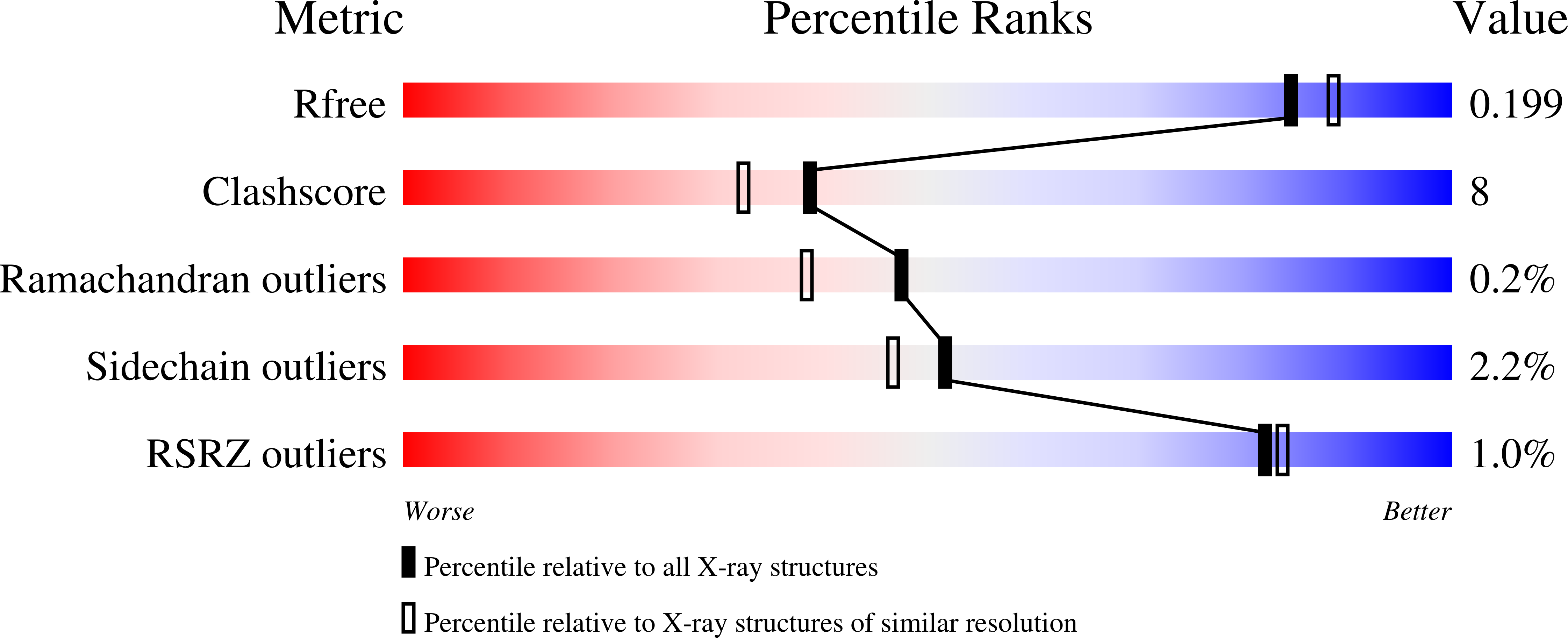
The structure of the neutral peroxidase from Arabidopsis thaliana (ATP N) has been determined to a resolution of 1.9 A and a free R value of 20.5%. ATP N has the expected characteristic fold of the class III peroxidases, with a C(alpha) r.m.s.d. of 0.82 A when compared with horseradish peroxidase C (HRP C). HRP C is 54% identical to ATP N in sequence. When the structures of four class III plant peroxidases are superimposed, the regions with structural differences are non-randomly distributed; all are located in one half of the molecule. The architecture of the haem pocket of ATP N is very similar to that of HRP C, in agreement with the low small-molecule substrate specificity of all class III peroxidases. The structure of ATP N suggests that the pH dependence of the substrate turnover will differ from that of HRP C owing to differences in polarity of the residues in the substrate-access channel. Since there are fewer hydrogen bonds to haem C17 propionate O atoms in ATP N than in HRP C, it is suggested that ATP N will lose haem more easily than HRP C. Unlike almost all other class III plant peroxidases, ATP N has a free cysteine residue at a similar position to the suggested secondary substrate-binding site in lignin peroxidase.
Organizational Affiliation:
Protein Structure Group, Department of Chemistry, University of Copenhagen, Universitetsparken 5, DK-2100 Kobenhavn O, Denmark.