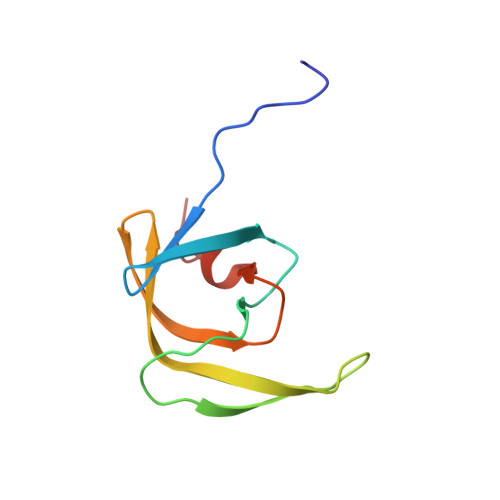
Solution structure of the mature HIV-1 protease monomer: Insight into the tertiary fold and stability of a precursor
Ishima, R., Torchia, D.A., Lynch, S.M., Gronenborn, A.M., Louis, J.M.(2003) J Biol Chem 278: 43311-43319
- PubMed: 12933791
- DOI: https://doi.org/10.1074/jbc.M307549200
- Primary Citation of Related Structures:
1Q9P - PubMed Abstract:
We present the first solution structure of the HIV-1 protease monomer spanning the region Phe1-Ala95 (PR1-95). Except for the terminal regions (residues 1-10 and 91-95) that are disordered, the tertiary fold of the remainder of the protease is essentially identical to that of the individual subunit of the dimer. In the monomer, the side chains of buried residues stabilizing the active site interface in the dimer, such as Asp25, Asp29, and Arg87, are now exposed to solvent. The flap dynamics in the monomer are similar to that of the free protease dimer. We also show that the protease domain of an optimized precursor flanked by 56 amino acids of the N-terminal transframe region is predominantly monomeric, exhibiting a tertiary fold that is quite similar to that of PR1-95 structure. This explains the very low catalytic activity observed for the protease prior to its maturation at its N terminus as compared with the mature protease, which is an active stable dimer under identical conditions. Adding as few as 2 amino acids to the N terminus of the mature protease significantly increases its dissociation into monomers. Knowledge of the protease monomer structure and critical features of its dimerization may aid in the screening and design of compounds that target the protease prior to its maturation from the Gag-Pol precursor.
Organizational Affiliation:
Molecular Structural Biology Unit, NIDCR, National Institutes of Health, Bethesda, Maryland 20892-4307, USA.