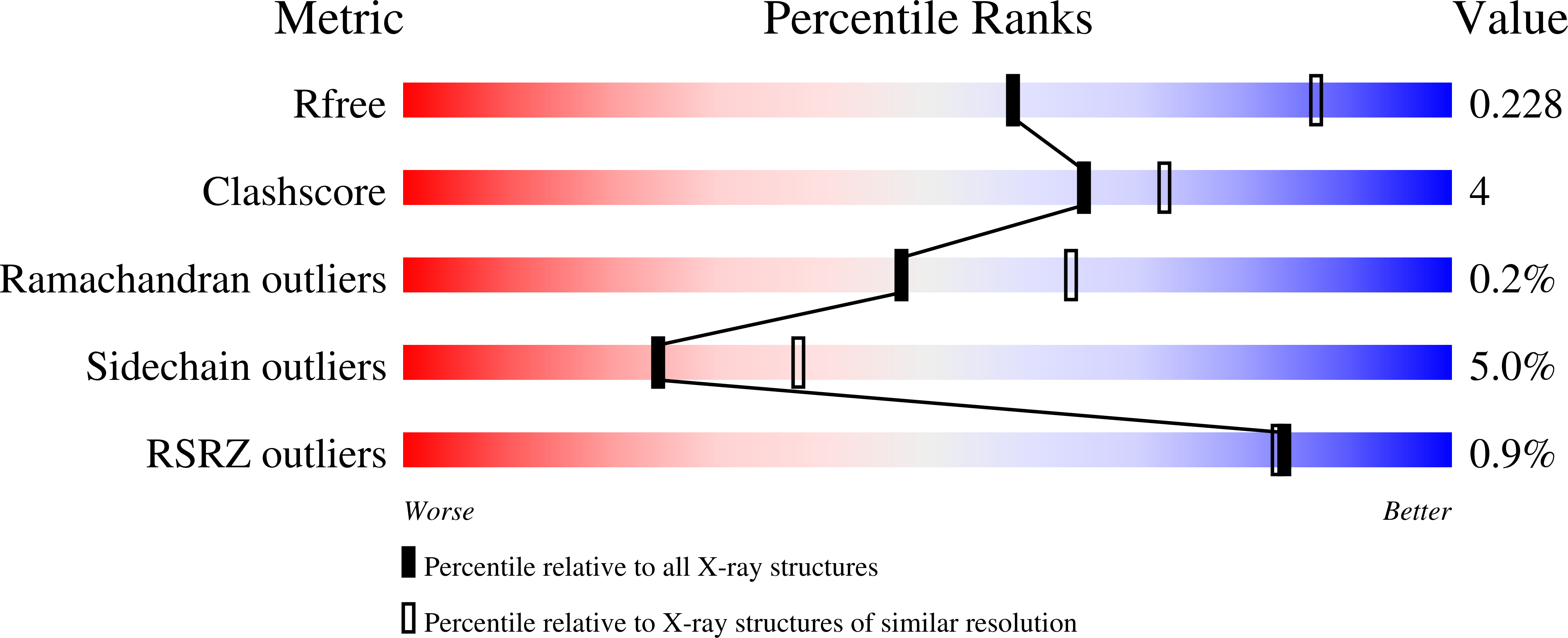
Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation.
Bourne, Y., Kolb, H.C., Radic, Z., Sharpless, K.B., Taylor, P., Marchot, P.(2004) Proc Natl Acad Sci U S A 101: 1449-1454
- PubMed: 14757816
- DOI: https://doi.org/10.1073/pnas.0308206100
- Primary Citation of Related Structures:
1Q83, 1Q84 - PubMed Abstract:
The 1,3-dipolar cycloaddition reaction between unactivated azides and acetylenes proceeds exceedingly slowly at room temperature. However, considerable rate acceleration is observed when this reaction occurs inside the active center gorge of acetylcholinesterase (AChE) between certain azide and acetylene reactants, attached via methylene chains to specific inhibitor moieties selective for the active center and peripheral site of the enzyme. AChE catalyzes the formation of its own inhibitor in a highly selective fashion: only a single syn1-triazole regioisomer with defined substitution positions and linker distances is generated from a series of reagent combinations. Inhibition measurements revealed this syn1-triazole isomer to be the highest affinity reversible organic inhibitor of AChE with association rate constants near the diffusion limit. The corresponding anti1 isomer, not formed by the enzyme, proved to be a respectable but weaker inhibitor. The crystal structures of the syn1- and anti1-mouse AChE complexes at 2.45- to 2.65-A resolution reveal not only substantial binding contributions from the triazole moieties, but also that binding of the syn1 isomer induces large and unprecedented enzyme conformational changes not observed in the anti1 complex nor predicted from structures of the apoenzyme and complexes with the precursor reactants. Hence, the freeze-frame reaction offers both a strategically original approach for drug discovery and a means for kinetically controlled capture, as a high-affinity complex between the enzyme and its self-created inhibitor, of a highly reactive minor abundance conformer of a fluctuating protein template.
Organizational Affiliation:
Ingénierie des Protéines, Centre National de la Recherche Scientifique Unité Mixte de Recherche-6560, Institut Fédératif de Recherche Jean Roche, Université de la Méditerranée, Faculté de Médecine Secteur Nord, F-13916 Marseille Cedex 20, France.