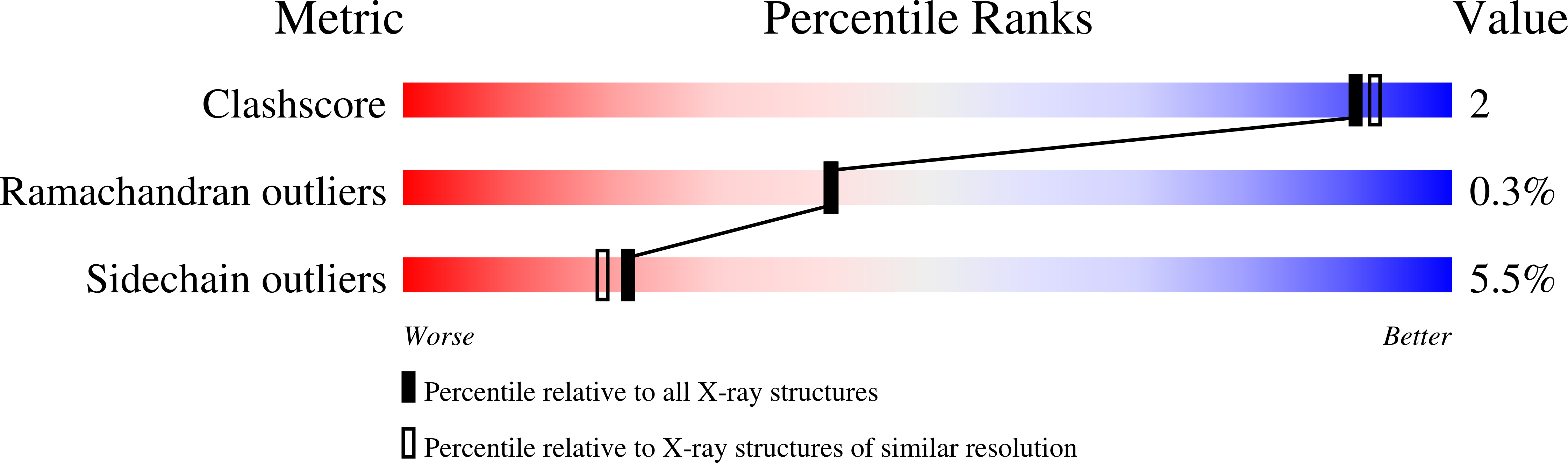Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Lah, M.S., Palfey, B.A., Schreuder, H.A., Ludwig, M.L.(1994) Biochemistry 33: 1555-1564
- PubMed: 8312276
- DOI: https://doi.org/10.1021/bi00172a036
- Primary Citation of Related Structures:
1PXA, 1PXB, 1PXC - PubMed Abstract:
Structures of the mutant p-hydroxybenzoate hydroxylases, Tyr201Phe, Tyr385Phe, and Asn300Asp, each complexed with the substrate p-OHB have been determined by X-ray crystallography. Crystals of these three mutants of the Pseudomonas aeruginosa enzyme, which differs from the wild-type Pseudomonas fluorescens enzyme at two surface positions (228 and 249), were isomorphous with crystals of the wild-type P. fluorescens enzyme, allowing the mutant structures to be determined by model building and refinement, starting from the coordinates for the oxidized P. fluorescens PHBH-3,4-diOHB complex [Schreuder, H.A., van der Laan, J.M., Hol, W.G.J., & Drenth, J. (1988) J. Mol. Biol. 199, 637-648]. The R factors for the structures described here are: Tyr385Phe, 0.178 for data from 40.0 to 2.1 A; Tyr201Phe, 0.203 for data from 40.0 to 2.3 A; and Asn300Asp, 0.193 for data from 40.0 to 2.3 A. The functional effects of the Tyr201Phe and Tyr385Phe mutations, described earlier [Entsch, B., Palfey, B.A., Ballou, D.P., & Massey, V. (1991) J. Biol. Chem. 266, 17341-17349], were rationalized with the assumption that the mutations perturbed the hydrogen-bonding interactions of the tyrosine residues but caused no other changes in the enzyme structure. In agreement with these assumptions, the positions of the substrate, the flavin, and the modified residues are not altered in the Tyr385Phe and Tyr201Phe structures. In contrast, substitution of Asp for Asn at residue 300 has more profound effects on the enzyme structure. The side chain of Asp300 moves away from the flavin, disrupting the interactions of the carboxamide group with the flavin O(2) atom, and the alpha-helix H10 that begins at residue 297 is displaced, altering its dipole interactions with the flavin ring. The functional consequences of these changes in the enzyme structure and of the introduction of the carboxyl group at 300 are described and discussed in the accompanying paper (Palfey et al., 1994b).
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, Ann Arbor 48109.