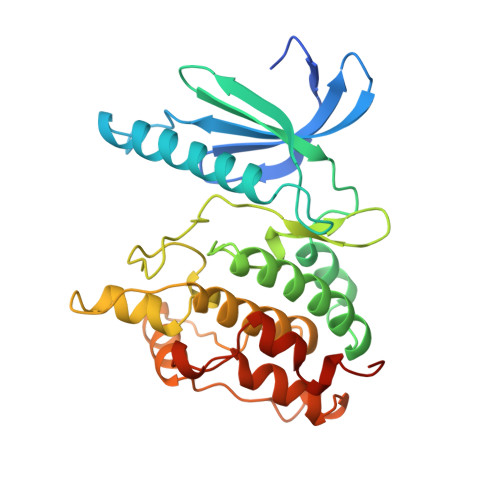
Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
Owen, D.J., Noble, M.E., Garman, E.F., Papageorgiou, A.C., Johnson, L.N.(1995) Structure 3: 467-482
- PubMed: 7663944
- DOI: https://doi.org/10.1016/s0969-2126(01)00180-0
- Primary Citation of Related Structures:
1PHK - PubMed Abstract:
Control of intracellular events by protein phosphorylation is promoted by specific protein kinases. All the known protein kinase possess a common structure that defines a catalytically competent entity termed the 'kinase catalytic core'. Within this common structural framework each kinase displays its own unique substrate specificity, and a regulatory mechanism that may be modulated by association with other proteins. Structural studies of phosphorylase kinase (Phk), the major substrate of which is glycogen phosphorylase, may be expected to shed light on its regulation.
Organizational Affiliation:
Laboratory of Molecular Biophysics, University of Oxford, UK.