Solution structure of the single-stranded DNA binding protein of the filamentous Pseudomonas phage Pf3: similarity to other proteins binding to single-stranded nucleic acids.
Folmer, R.H., Nilges, M., Konings, R.N., Hilbers, C.W.(1995) EMBO J 14: 4132-4142
- PubMed: 7556054
- DOI: https://doi.org/10.1002/j.1460-2075.1995.tb00087.x
- Primary Citation of Related Structures:
1PFS - PubMed Abstract:
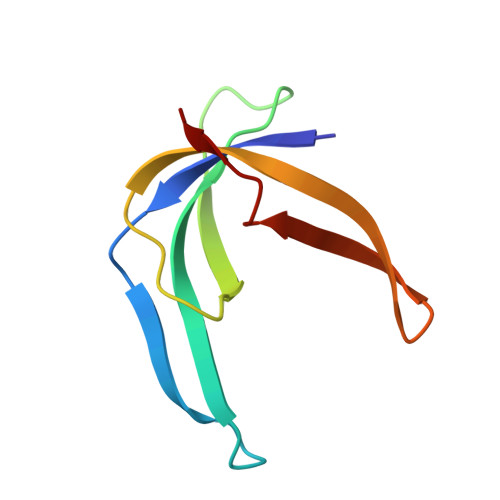
The three-dimensional structure of the homodimeric single-stranded DNA binding protein encoded by the filamentous Pseudomonas bacteriophage Pf3 has been determined using heteronuclear multidimensional NMR techniques and restrained molecular dynamics. NMR experiments and structure calculations have been performed on a mutant protein (Phe36 --> His) that was successfully designed to reduce the tendency of the protein to aggregate. The protein monomer is composed of a five-stranded antiparallel beta-sheet from which two beta-hairpins and a large loop protrude. The structure is compared with the single-stranded DNA binding protein encoded by the filamentous Escherichia coli phage Ff, a protein with a similar biological function and DNA binding properties, yet quite different amino acid sequence, and with the major cold shock protein of Escherichia coli, a single-stranded DNA binding protein with an entirely different sequence, biological function and binding characteristics. The amino acid sequence of the latter is highly homologous to the nucleic acid binding domain (i.e. the cold shock domain) of proteins belonging to the Y-box family. Despite their differences in amino acid sequence and function, the folds of the three proteins are remarkably similar, suggesting that this is a preferred folding pattern shared by many single-stranded DNA binding proteins.
Organizational Affiliation:
Nijmegen SON Research Center, University of Nijmegen, The Netherlands.