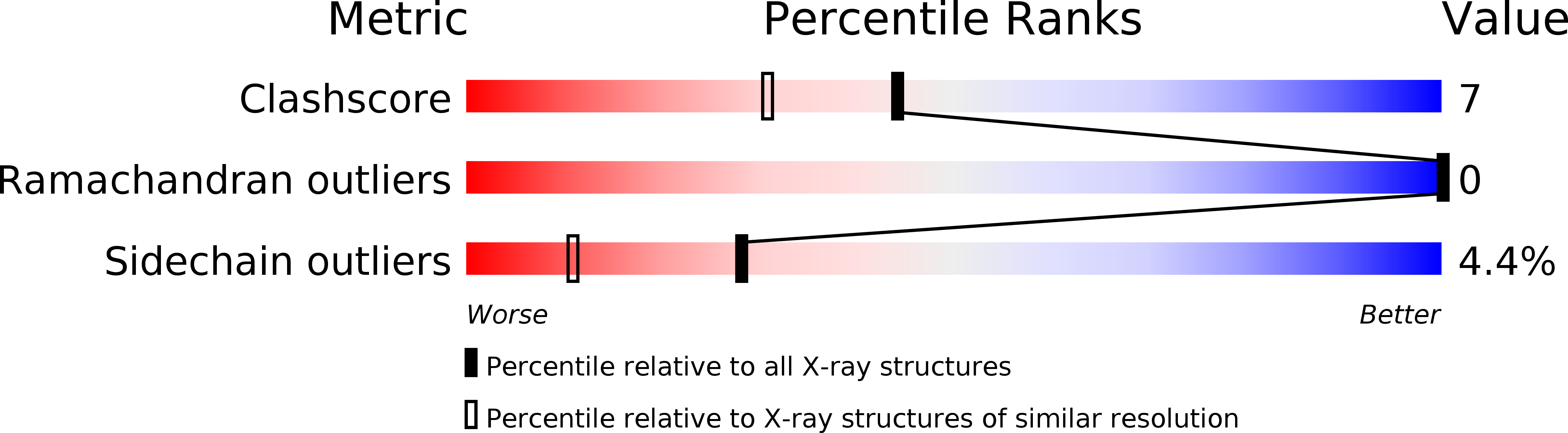Structural Biology: A New Model for Protein Stereospecificity.
Mesecar, A.D., Koshland Jr., D.E.(2000) Nature 403: 614-615
- PubMed: 10688187
- DOI: https://doi.org/10.1038/35001144
- Primary Citation of Related Structures:
1P8F, 1PB1
Organizational Affiliation:
Center for Pharmaceutical Biotechnology, Department of Medicinal Chemistry and Pharmacognosy, University of Illinois at Chicago, 60607, USA.