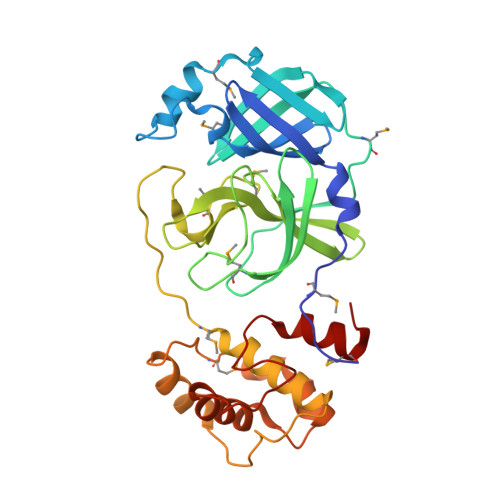
Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J.R., Hilgenfeld, R.(2003) Science 300: 1763-1767
- PubMed: 12746549
- DOI: https://doi.org/10.1126/science.1085658
- Primary Citation of Related Structures:
1P9S, 1P9U - PubMed Abstract:
A novel coronavirus has been identified as the causative agent of severe acute respiratory syndrome (SARS). The viral main proteinase (Mpro, also called 3CLpro), which controls the activities of the coronavirus replication complex, is an attractive target for therapy. We determined crystal structures for human coronavirus (strain 229E) Mpro and for an inhibitor complex of porcine coronavirus [transmissible gastroenteritis virus (TGEV)] Mpro, and we constructed a homology model for SARS coronavirus (SARS-CoV) Mpro. The structures reveal a remarkable degree of conservation of the substrate-binding sites, which is further supported by recombinant SARS-CoV Mpro-mediated cleavage of a TGEV Mpro substrate. Molecular modeling suggests that available rhinovirus 3Cpro inhibitors may be modified to make them useful for treating SARS.
Organizational Affiliation:
Institute of Biochemistry, University of Lübeck, D-23538 Lübeck, Germany.