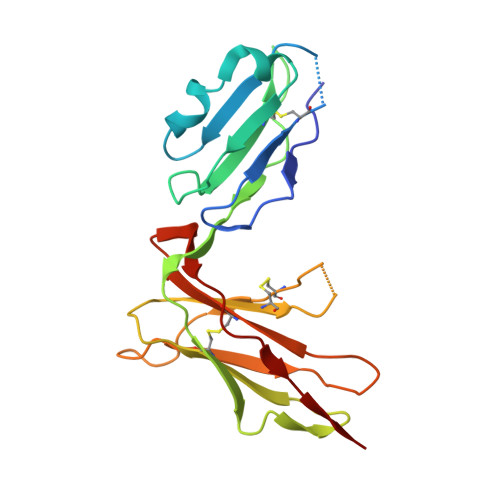
Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor.
Willcox, B.E., Thomas, L.M., Bjorkman, P.J.(2003) Nat Immunol 4: 913-919
- PubMed: 12897781
- DOI: https://doi.org/10.1038/ni961
- Primary Citation of Related Structures:
1P7Q - PubMed Abstract:
Leukocyte immunoglobulin-like receptor 1 (LIR-1), an inhibitory receptor expressed on monocytes, dendritic cells and lymphocytes, regulates cellular function by binding a broad range of classical and nonclassical major histocompatibility complex (MHC) class I molecules, and the human cytomegalovirus MHC class I homolog UL18. Here we describe the 3.4-A crystal structure of a complex between the LIR-1 D1D2 domains and the MHC class I molecule HLA-A2. LIR-1 contacts the mostly conserved beta(2)-microglobulin and alpha3 domains of HLA-A2. The LIR-1 binding site comprises residues at the interdomain hinge, and a patch at the D1 tip. The structure shows how LIR-1 recognizes UL18 and diverse MHC class I molecules, and indicates that a similar mode of MHC class I recognition is used by other LIR family members.
Organizational Affiliation:
Division of Biology 114-96, California Institute of Technology, Pasadena, California 91125, USA.