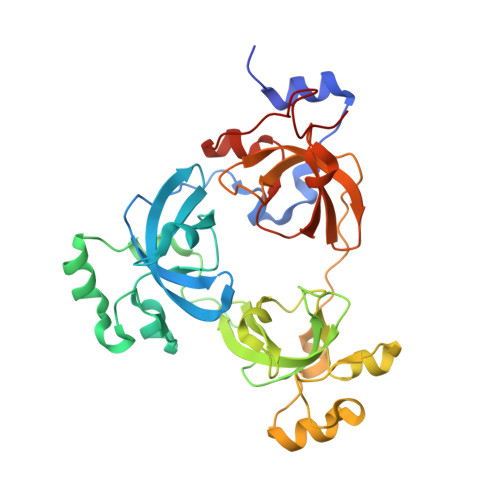
Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Wang, W.K., Tereshko, V., Boccuni, P., MacGrogan, D., Nimer, S.D., Patel, D.J.(2003) Structure 11: 775-789
- PubMed: 12842041
- DOI: https://doi.org/10.1016/s0969-2126(03)00127-8
- Primary Citation of Related Structures:
1OYX, 1OZ2, 1OZ3 - PubMed Abstract:
We report on the X-ray structure of three 100-amino acid mbt repeats in h-l(3)mbt, a polycomb group protein involved in transcriptional repression, whose gene is located in a region of chromosome 20 associated with hematopoietic malignancies. Interdigitation between the extended arms and cores of the mbt repeats results in a three-leaved propeller-like architecture, containing a central cavity. We have identified one ligand binding pocket per mbt repeat, which accommodates either the morphilino ring of MES or the proline ring of the C-terminal peptide segment, within a cavity lined by aromatic amino acids. Strikingly, phenotypic alterations resulting from point mutations or deletions in the mbt repeats of the related Drosophila SCM protein are clustered in and around the ligand binding pocket.
Organizational Affiliation:
Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10021, USA.