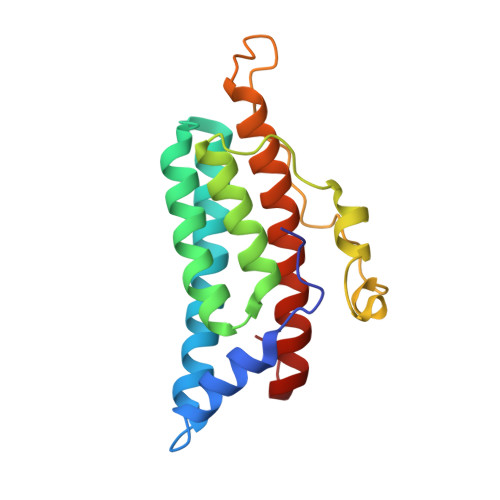
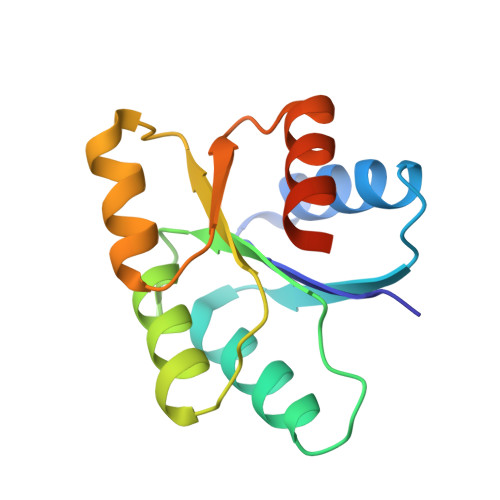
The yeast YPD1/SLN1 complex: insights into molecular recognition in two-component signaling systems.
Xu, Q., Porter, S.W., West, A.H.(2003) Structure 11: 1569-1581
- PubMed: 14656441
- DOI: https://doi.org/10.1016/j.str.2003.10.016
- Primary Citation of Related Structures:
1OXB, 1OXK - PubMed Abstract:
In Saccharomyces cerevisiae, a branched multistep phosphorelay signaling pathway regulates cellular adaptation to hyperosmotic stress. YPD1 functions as a histidine-phosphorylated protein intermediate required for phosphoryl group transfer from a membrane-bound sensor histidine kinase (SLN1) to two distinct response regulator proteins (SSK1 and SKN7). These four proteins are evolutionarily related to the well-characterized "two-component" regulatory proteins from bacteria. Although structural information is available for many two-component signaling proteins, there are very few examples of complexes between interacting phosphorelay partners. Here we report the first crystal structure of a prototypical monomeric histidine-containing phosphotransfer (HPt) protein YPD1 in complex with its upstream phosphodonor, the response regulator domain associated with SLN1.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Oklahoma, 620 Parrington Oval, Norman, OK 73019, USA.