DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site.
Li, C.-L., Hor, L.-I., Chang, Z.-F., Tsai, L.-C., Yang, W.-Z., Yuan, H.S.(2003) EMBO J 22: 4014-4025
- PubMed: 12881435
- DOI: https://doi.org/10.1093/emboj/cdg377
- Primary Citation of Related Structures:
1OUO, 1OUP - PubMed Abstract:
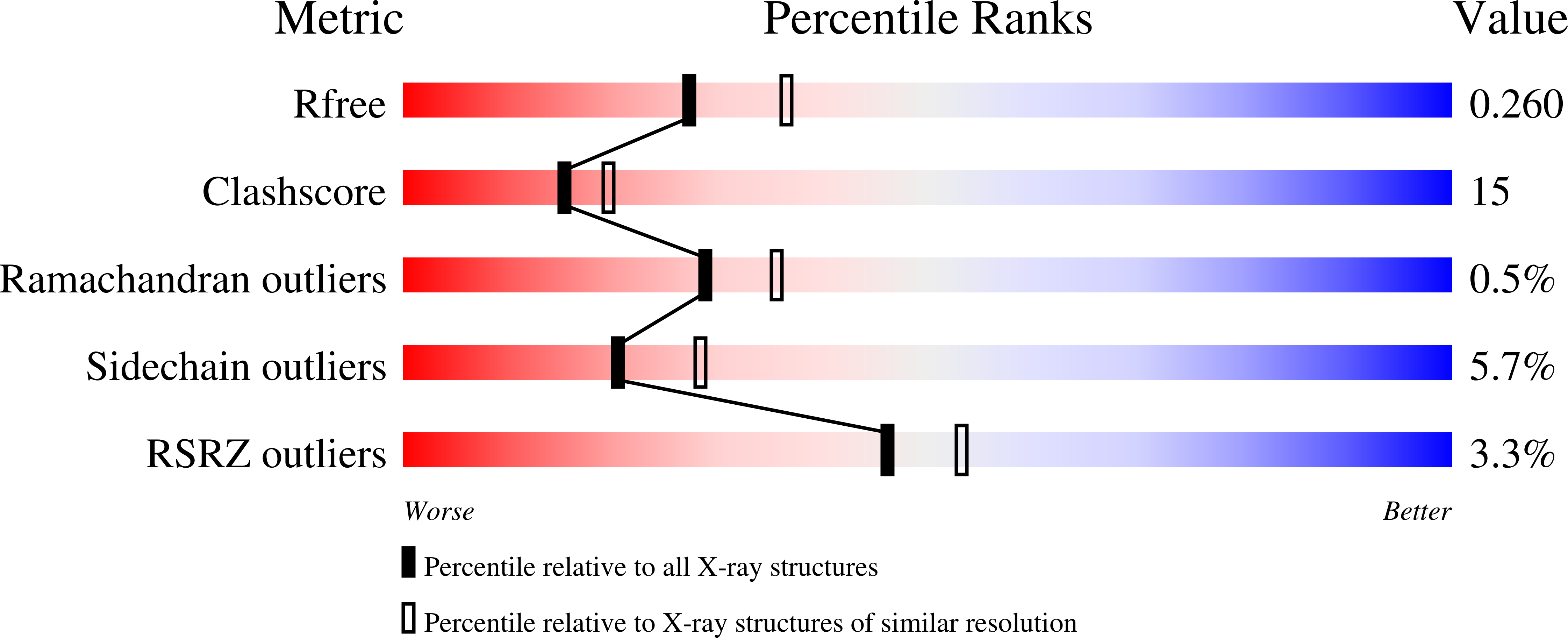
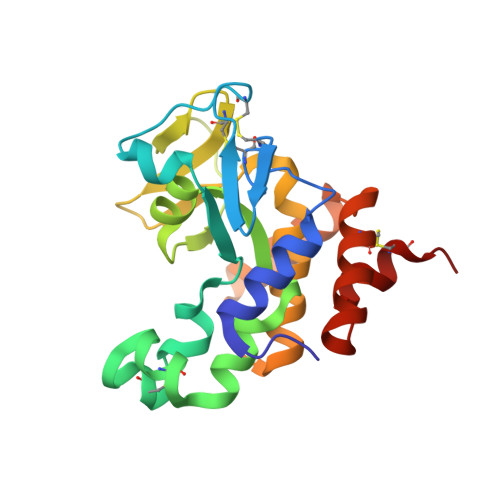
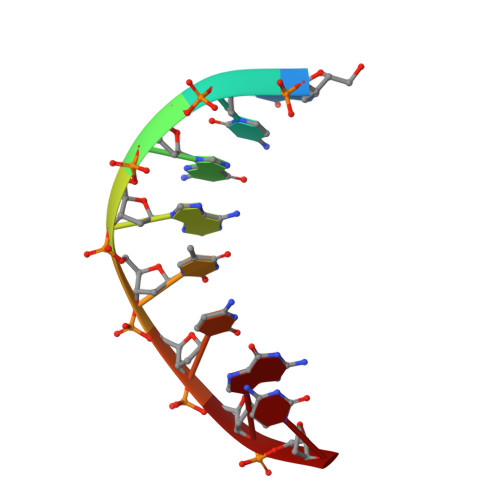
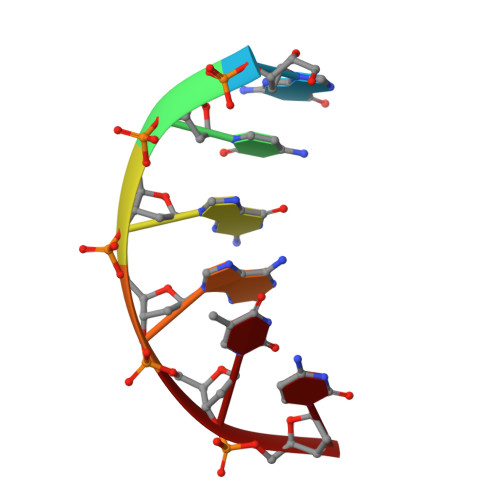
The Vibrio vulnificus nuclease, Vvn, is a non-specific periplasmic nuclease capable of digesting DNA and RNA. The crystal structure of Vvn and that of Vvn mutant H80A in complex with DNA were resolved at 2.3 A resolution. Vvn has a novel mixed alpha/beta topology containing four disulfide bridges, suggesting that Vvn is not active under reducing conditions in the cytoplasm. The overall structure of Vvn shows no similarity to other endonucleases; however, a known 'betabetaalpha-metal' motif is identified in the central cleft region. The crystal structure of the mutant Vvn-DNA complex demonstrates that Vvn binds mainly at the minor groove of DNA, resulting in duplex bending towards the major groove by approximately 20 degrees. Only the DNA phosphate backbones make hydrogen bonds with Vvn, suggesting a structural basis for its sequence-independent recognition of DNA and RNA. Based on the enzyme-substrate and enzyme-product structures observed in the mutant Vvn-DNA crystals, a catalytic mechanism is proposed. This structural study suggests that Vvn hydrolyzes DNA by a general single-metal ion mechanism, and indicates how non-specific DNA-binding proteins may recognize DNA.
Organizational Affiliation:
Institute of Biochemistry and Molecular Biology, College of Medicine, National Taiwan University, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan, Republic of China.