Crystal structure at 3 A of mistletoe lectin I, a dimeric type-II ribosome-inactivating protein, complexed with galactose
Niwa, H., Tonevitsky, A.G., Agapov, I.I., Saward, S., Pfuller, U., Palmer, R.A.(2003) Eur J Biochem 270: 2739-2749
- PubMed: 12823544
- DOI: https://doi.org/10.1046/j.1432-1033.2003.03646.x
- Primary Citation of Related Structures:
1OQL - PubMed Abstract:
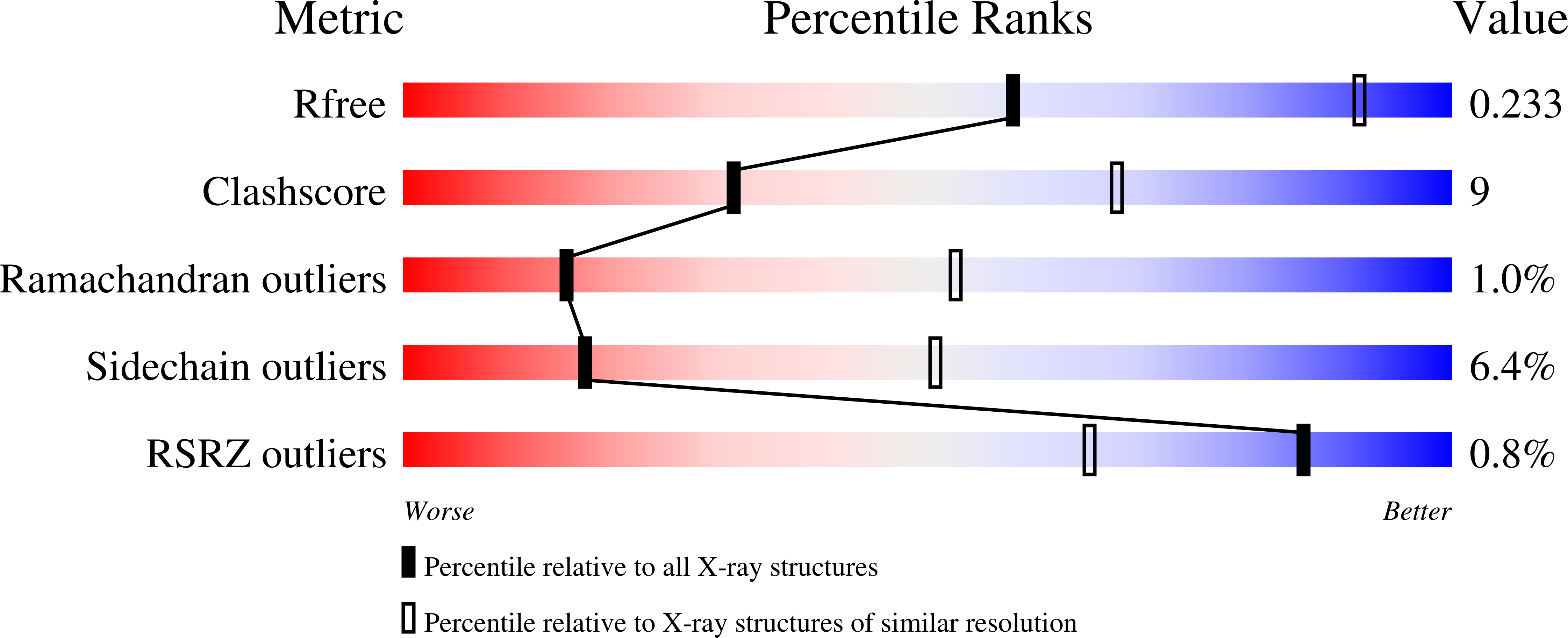
The X-ray structure of mistletoe lectin I (MLI), a type-II ribosome-inactivating protein (RIP), cocrystallized with galactose is described. The model was refined at 3.0 A resolution to an R-factor of 19.9% using 21 899 reflections, with Rfree 24.0%. MLI forms a homodimer (A-B)2 in the crystal, as it does in solution at high concentration. The dimer is formed through contacts between the N-terminal domains of two B-chains involving weak polar and non-polar interactions. Consequently, the overall arrangement of sugar-binding sites in MLI differs from those in monomeric type-II RIPs: two N-terminal sugar-binding sites are 15 A apart on one side of the dimer, and two C-terminal sugar-binding sites are 87 A apart on the other side. Galactose binding is achieved by common hydrogen bonds for the two binding sites via hydroxy groups 3-OH and 4-OH and hydrophobic contact by an aromatic ring. In addition, at the N-terminal site 2-OH forms hydrogen bonds with Asp27 and Lys41, and at the C-terminal site 3-OH and 6-OH undergo water-mediated interactions and C5 has a hydrophobic contact. MLI is a galactose-specific lectin and shows little affinity for N-acetylgalactosamine. The reason for this is discussed. Structural differences among the RIPs investigated in this study (their quaternary structures, location of sugar-binding sites, and fine sugar specificities of their B-chains, which could have diverged through evolution from a two-domain protein) may affect the binding sites, and consequently the cellular transport processes and biological responses of these toxins.
Organizational Affiliation:
School of Crystallography, Birkbeck College, University of London, Malet Street, London WC1E 7HX, UK. h.niwa@mail.cryst.bbk.ac.uk