Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Lukacik, P., Roversi, P., White, J., Esser, D., Smith, G.P., Billington, J., Williams, P.A., Rudd, P.M., Wormald, M.R., Harvey, D.J., Crispin, M.D.M., Radcliffe, C.M., Dwek, R.A., Evans, D.J., Morgan, B.P., Smith, R.A.G., Lea, S.M.(2004) Proc Natl Acad Sci U S A 101: 1279
- PubMed: 14734808
- DOI: https://doi.org/10.1073/pnas.0307200101
- Primary Citation of Related Structures:
1OJV, 1OJW, 1OJY, 1OK1, 1OK2, 1OK3, 1OK9 - PubMed Abstract:
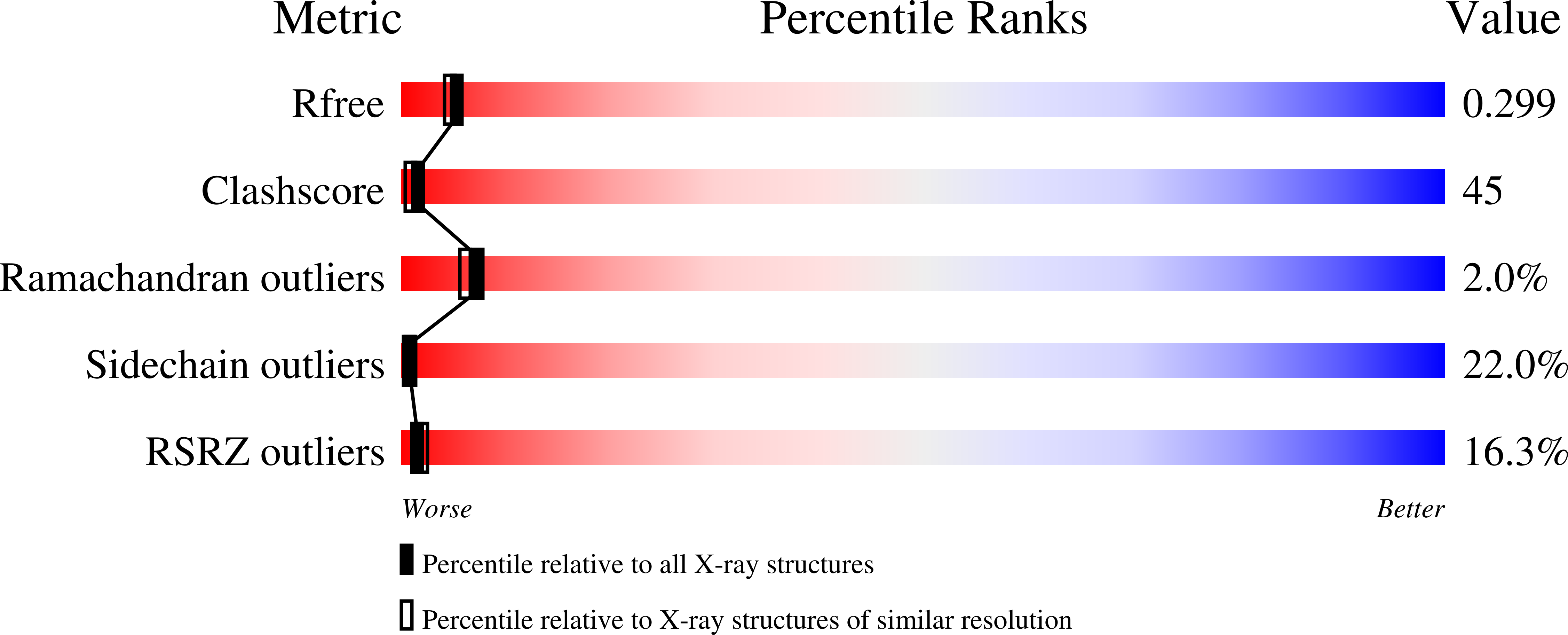
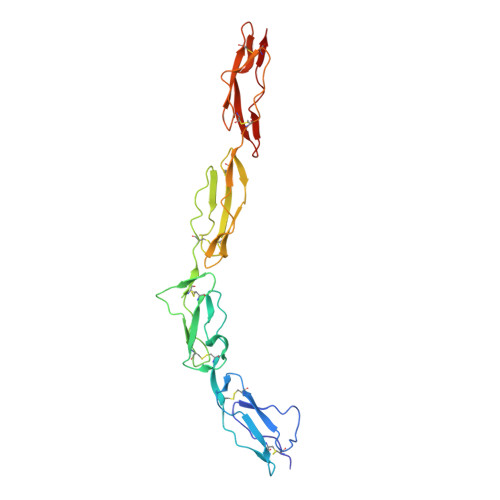
The human complement regulator CD55 is a key molecule protecting self-cells from complement-mediated lysis. X-ray diffraction and analytical ultracentrifugation data reveal a rod-like arrangement of four short consensus repeat (SCR) domains in both the crystal and solution. The stalk linking the four SCR domains to the glycosylphosphatidylinositol anchor is extended by the addition of 11 highly charged O-glycans and positions the domains an estimated 177 A above the membrane. Mutation mapping and hydrophobic potential analysis suggest that the interaction with the convertase, and thus complement regulation, depends on the burial of a hydrophobic patch centered on the linker between SCR domains 2 and 3.
Organizational Affiliation:
Laboratory of Molecular Biophysics and Glycobiology Institute, Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, England.