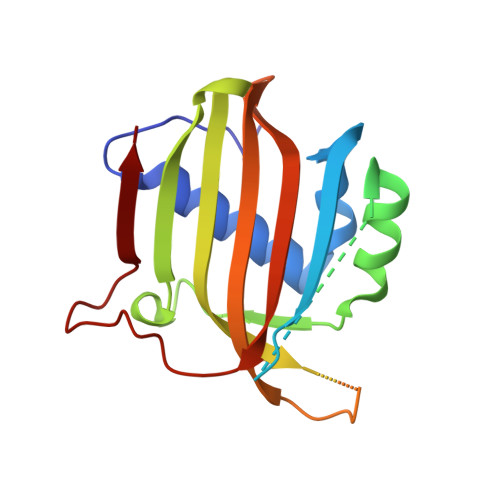
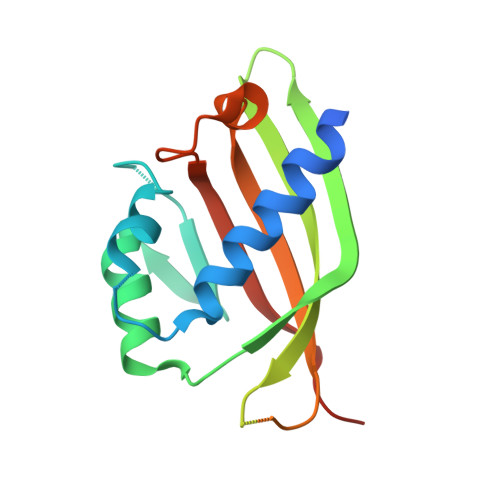
Structural similarity in the absence of sequence homology of the messenger RNA export factors Mtr2 and p15.
Fribourg, S., Conti, E.(2003) EMBO Rep 4: 699-703
- PubMed: 12835756
- DOI: https://doi.org/10.1038/sj.embor.embor883
- Primary Citation of Related Structures:
1OF5 - PubMed Abstract:
The association between Mtr2 and Mex67 is essential for the nuclear export of bulk messenger RNA in yeast. In metazoans, the analogous function is carried out by the TAP-p15 heterodimer. Whereas Mex67 and TAP are highly conserved proteins, their binding partners, Mtr2 and p15, share no sequence similarity, but are nevertheless functionally homologous. Here, we report the 2.8-A resolution crystal structure of Mtr2 in complex with the NTF2-like domain of Mex67. Mtr2 is a novel member of the NTF2-like family and interacts with Mex67, forming a complex with a similar structural architecture to that of TAP-p15. Mtr2 fulfils an analogous function to that of human p15 in maintaining the structural integrity of the heterodimer. In addition, Mtr2 presents a long internal loop, which contains residues that affect the export of the large ribosomal subunit.
Organizational Affiliation:
European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117 Heidelberg, Germany.