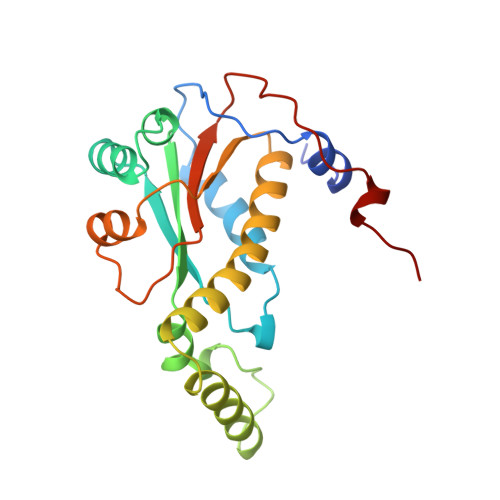
Crystal structure of NADH oxidase from Thermus thermophilus.
Hecht, H.J., Erdmann, H., Park, H.J., Sprinzl, M., Schmid, R.D.(1995) Nat Struct Biol 2: 1109-1114
- PubMed: 8846223
- DOI: https://doi.org/10.1038/nsb1295-1109
- Primary Citation of Related Structures:
1NOX - PubMed Abstract:
The crystal structures of the flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) containing isoforms of NADH oxidase from Thermus thermophilus have been determined by isomorphous and molecular replacement and refined to 2.3 A and 1.6 A resolution with R-values of 18.5% and 18.6% respectively. The structure of the homodimeric enzyme consists of a central 4-stranded antiparallel beta-sheet covered by helices, a more flexible domain formed by two helices, and a C-terminal excursion connecting the subunits. The active sites are located in a deep cleft between the subunits. The binding site of the flavin cofactor lacks the common nucleotide binding fold and is different from the FMN binding site found in flavodoxins.
Organizational Affiliation:
Department of Molecular Structure Research, GBF (Gesellschaft fr Biotechnologische, Forschung, Braunschweig, Germany.