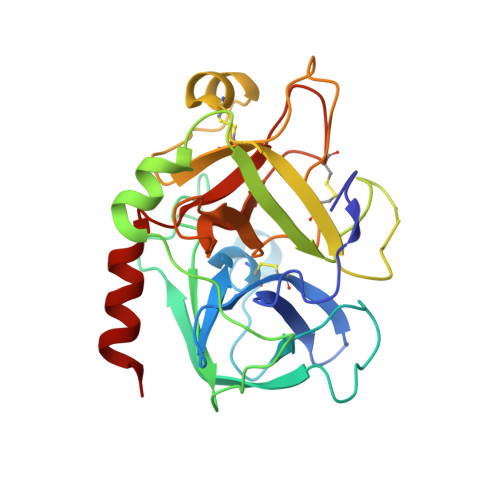
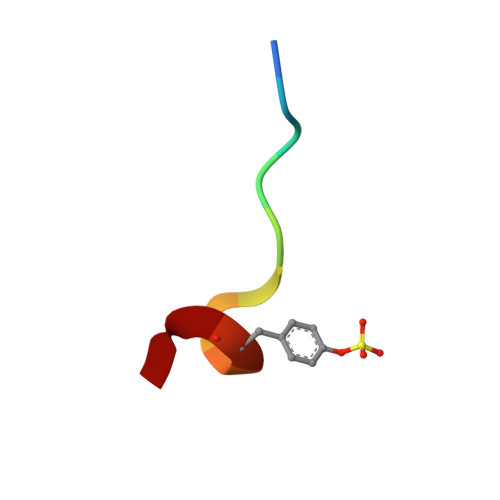
DESIGN OF WEAKLY BASIC THROMBIN INHIBITORS INCORPORATING NOVEL P1 BINDING FUNCTIONS: MOLECULAR AND X-RAY CRYSTALLOGRAPHIC STUDIES
De Simone, G., Menchise, V., Omaggio, S., Pedone, C., Scozzafava, A., Supuran, C.T.(2003) Biochemistry 42: 9013-9021
- PubMed: 12885234
- DOI: https://doi.org/10.1021/bi020512l
- Primary Citation of Related Structures:
1NO9 - PubMed Abstract:
To prepare weakly basic thrombin inhibitors with modified S1 anchoring groups, two series of compounds were synthesized by reaction of guanidine or aminoguanidine with acyl halides and N,N-disubstituted carbamoyl chlorides. pK(a) measurements of these acylated guanidines/aminoguanidines showed a reduced basicity, with pK(a) values in the range of 8.4-8.7. These molecules typically showed inhibition constants in the range of 150-425 nM against thrombin and 360-965 nM against trypsin, even though some bulky derivatives, such as N,N-diphenylcarbamoylguanidine/aminoguanidine and their congeners, showed much stronger thrombin inhibitory activity, with inhibition constants in the range of 24-42 nM. Unexpectedly, very long incubation times with both proteases revealed that aminoguanidine derivatives behaved as irreversible inhibitors. To assess the molecular basis responsible for the high affinity observed for these molecules toward thrombin, the crystal structure of the thrombin-hirugen-N,N-diphenylcarbamoylaminoguanidine complex has been solved at 1.90 A resolution. The structural analysis of the complex revealed an unexpected interaction mode with the protease, resulting in an N,N-diphenylcarbamoyl intermediate covalently bound to the catalytic serine as a consequence of its hydrolysis together with the release of the aminoguanidine moiety. Surprisingly, in this covalent adduct a phenyl group was found in the S1 specificity pocket, which usually recognizes positively charged residues. These findings provide new insights in the design of low basicity serine protease inhibitors.
Organizational Affiliation:
Laboratorio di Chimica Inorganica e Bioinorganica, Università degli Studi, via della Lastruccia 3, Rm 188, Polo Scientifico, 50019 Sesto Fiorentino, Florence, Italy. gmg@chemistry.unina.it