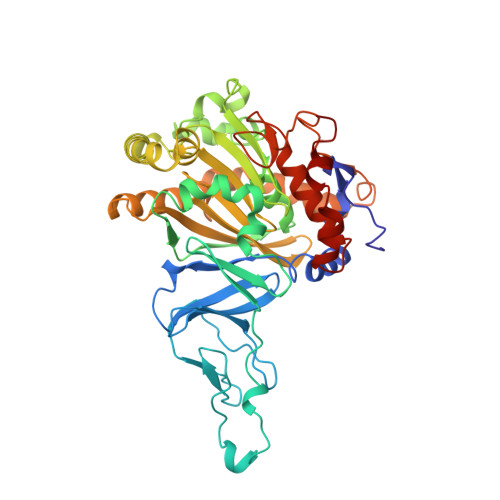
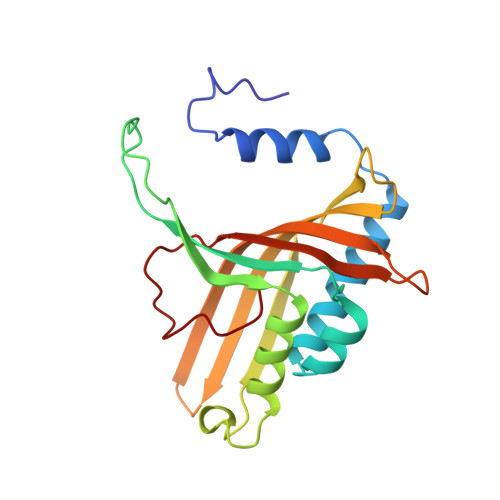
Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase.
Kauppi, B., Lee, K., Carredano, E., Parales, R.E., Gibson, D.T., Eklund, H., Ramaswamy, S.(1998) Structure 6: 571-586
- PubMed: 9634695
- DOI: https://doi.org/10.1016/s0969-2126(98)00059-8
- Primary Citation of Related Structures:
1NDO - PubMed Abstract:
Pseudomonas sp. NCIB 9816-4 utilizes a multicomponent enzyme system to oxidize naphthalene to (+)-cis-(1R,2S)-dihydroxy-1,2-dihydronaphthalene. The enzyme component catalyzing this reaction, naphthalene 1,2-dioxygenase (NDO), belongs to a family of aromatic-ring-hydroxylating dioxygenases that oxidize aromatic hydrocarbons and related compounds to cis-arene diols. These enzymes utilize a mononuclear non-heme iron center to catalyze the addition of dioxygen to their respective substrates. The present study was conducted to provide essential structural information necessary for elucidating the mechanism of action of NDO.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden.