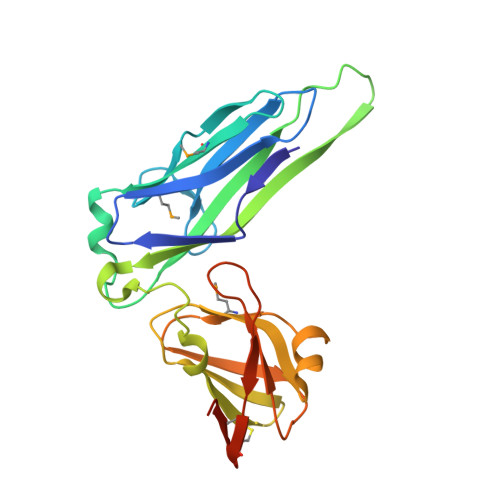
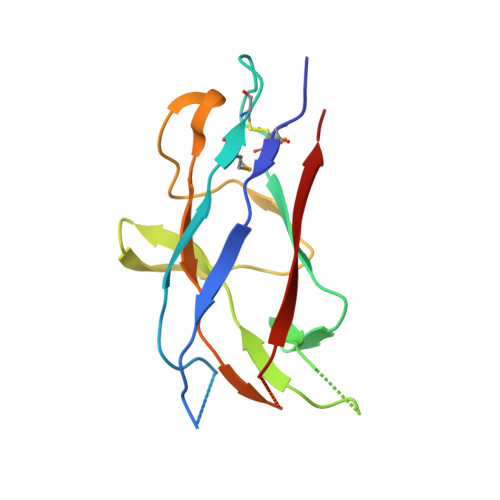
Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation
Sauer, F.G., Pinkner, J.S., Waksman, G., Hultgren, S.J.(2002) Cell 111: 543-551
- PubMed: 12437927
- DOI: https://doi.org/10.1016/s0092-8674(02)01050-4
- Primary Citation of Related Structures:
1N0L, 1N12 - PubMed Abstract:
Periplasmic chaperones direct the assembly of adhesive, multi-subunit pilus fibers that play critical roles in bacterial pathogenesis. Pilus assembly occurs via a donor strand exchange mechanism in which the N-terminal extension of one subunit replaces the chaperone G(1) strand that transiently occupies a groove in the neighboring subunit. Here, we show that the chaperone primes the subunit for assembly by holding the groove in an open, activated conformation. During donor strand exchange, the subunit undergoes a topological transition that triggers the closure of the groove and seals the N-terminal extension in place. It is this topological transition, made possible only by the priming action of the chaperone that drives subunit assembly into the fiber.
Organizational Affiliation:
Department of Molecular Microbiology, Washington University Medical School, 660 South Euclid Avenue, St. Louis, MO 63105, USA.