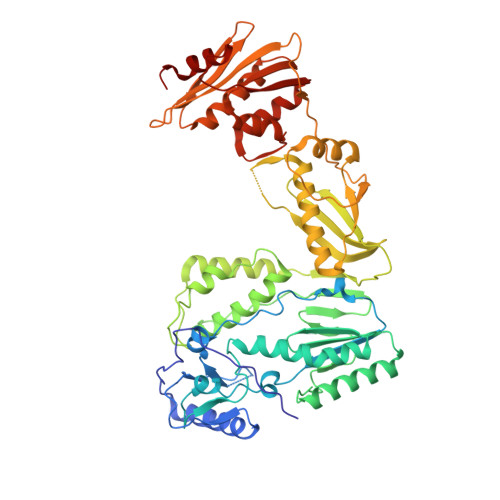
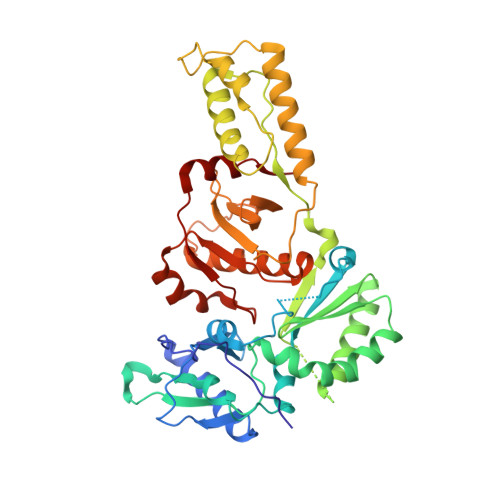
Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors
Ren, J., Bird, L.E., Chamberlain, P.P., Stewart-Jones, G.B., Stuart, D.I., Stammers, D.K.(2002) Proc Natl Acad Sci U S A 99: 14410-14415
- PubMed: 12386343
- DOI: https://doi.org/10.1073/pnas.222366699
- Primary Citation of Related Structures:
1MU2 - PubMed Abstract:
The HIV-2 serotype of HIV is a cause of disease in parts of the West African population, and there is evidence for its spread to Europe and Asia. HIV-2 reverse transcriptase (RT) demonstrates an intrinsic resistance to non-nucleoside RT inhibitors (NNRTIs), one of two classes of anti-AIDS drugs that target the viral RT. We report the crystal structure of HIV-2 RT to 2.35 A resolution, which reveals molecular details of the resistance to NNRTIs. HIV-2 RT has a similar overall fold to HIV-1 RT but has structural differences within the "NNRTI pocket" at both conserved and nonconserved residues. The structure points to the role of sequence differences that can give rise to unfavorable inhibitor contacts or destabilization of part of the binding pocket at positions 101, 106, 138, 181, 188, and 190. We also present evidence that the conformation of Ile-181 compared with the HIV-1 Tyr-181 could be a significant contributory factor to this inherent drug resistance of HIV-2 to NNRTIs. The availability of a refined structure of HIV-2 RT will provide a stimulus for the structure-based design of novel non-nucleoside inhibitors that could be used against HIV-2 infection.
Organizational Affiliation:
Division of Structural Biology, The Wellcome Trust Centre for Human Genetics, The Henry Wellcome Building for Genomic Medicine, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, United Kingdom.