Structural Basis for the Transition from Initiation to Elongation Transcription in T7 RNA Polymerase
Yin, Y.W., Steitz, T.A.(2002) Science 298: 1387-1395
- PubMed: 12242451
- DOI: https://doi.org/10.1126/science.1077464
- Primary Citation of Related Structures:
1MSW - PubMed Abstract:
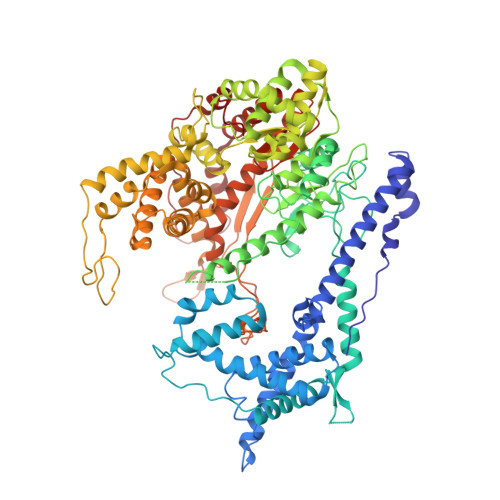
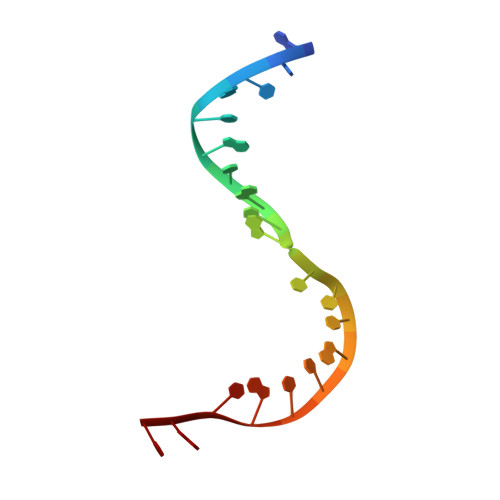
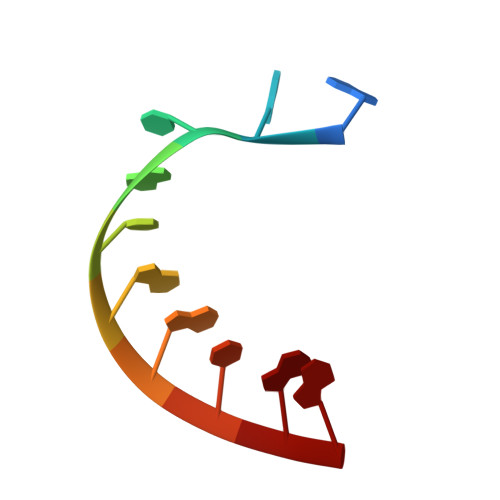
To make messenger RNA transcripts, bacteriophage T7 RNA polymerase (T7 RNAP) undergoes a transition from an initiation phase, which only makes short RNA fragments, to a stable elongation phase. We have determined at 2.1 angstrom resolution the crystal structure of a T7 RNAP elongation complex with 30 base pairs of duplex DNA containing a "transcription bubble" interacting with a 17-nucleotide RNA transcript. The transition from an initiation to an elongation complex is accompanied by a major refolding of the amino-terminal 300 residues. This results in loss of the promoter binding site, facilitating promoter clearance, and creates a tunnel that surrounds the RNA transcript after it peels off a seven-base pair heteroduplex. Formation of the exit tunnel explains the enhanced processivity of the elongation complex. Downstream duplex DNA binds to the fingers domain, and its orientation relative to upstream DNA in the initiation complex implies an unwinding that could facilitate formation of the open promoter complex.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, 266 Whitney Avenue, New Haven, CT 06520-8114, USA.