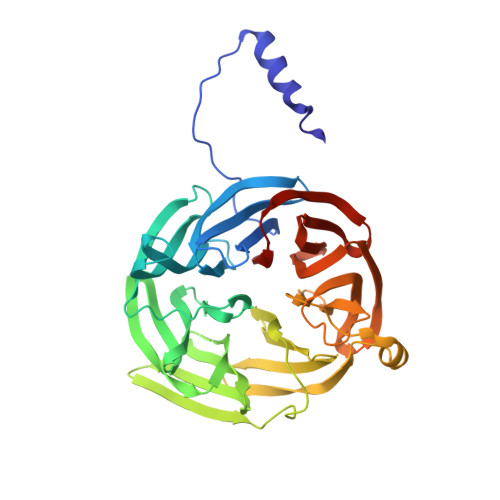
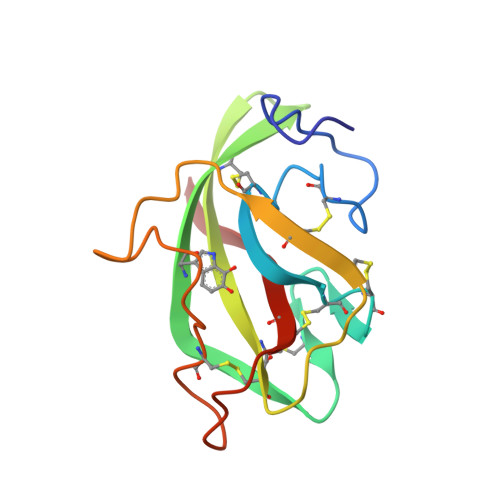
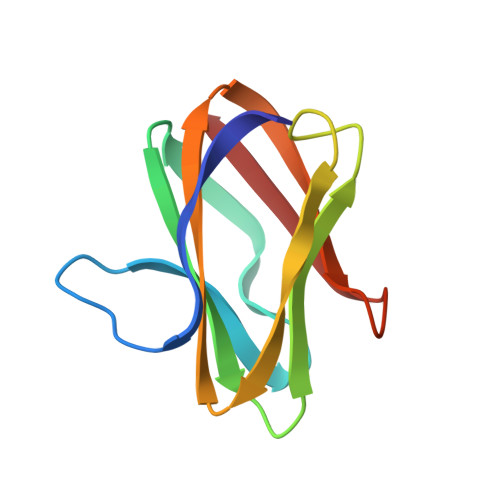
Crystal structure of an electron-transfer complex between methylamine dehydrogenase and amicyanin.
Chen, L., Durley, R., Poliks, B.J., Hamada, K., Chen, Z., Mathews, F.S., Davidson, V.L., Satow, Y., Huizinga, E., Vellieux, F.M., Hol, W.G.J.(1992) Biochemistry 31: 4959-4964
- PubMed: 1599920
- DOI: https://doi.org/10.1021/bi00136a006
- Primary Citation of Related Structures:
1MDA - PubMed Abstract:
The crystal structure of the complex between the quinoprotein methylamine dehydrogenase (MADH) and the type I blue copper protein amicyanin, both from Paracoccus denitrificans, has been determined at 2.5-A resolution using molecular replacement. The search model was MADH from Thiobacillus versutus. The amicyanin could be located in an averaged electron density difference map and the model improved by refinement and model building procedures. Nine beta-strands are observed within the amicyanin molecule. The copper atom is located between three antiparallel strands and is about 2.5 A below the protein surface. The major intermolecular interactions occur between amicyanin and the light subunit of MADH where the interface is largely hydrophobic. The copper atom of amicyanin and the redox cofactor of MADH are about 9.4 A apart. One of the copper ligands, His 95, lies between the two redox centers and may facilitate electron transfer between them.
Organizational Affiliation:
Department of Cell Biology and Physiology, Washington University School of Medicine, St. Louis, Missouri 63110.