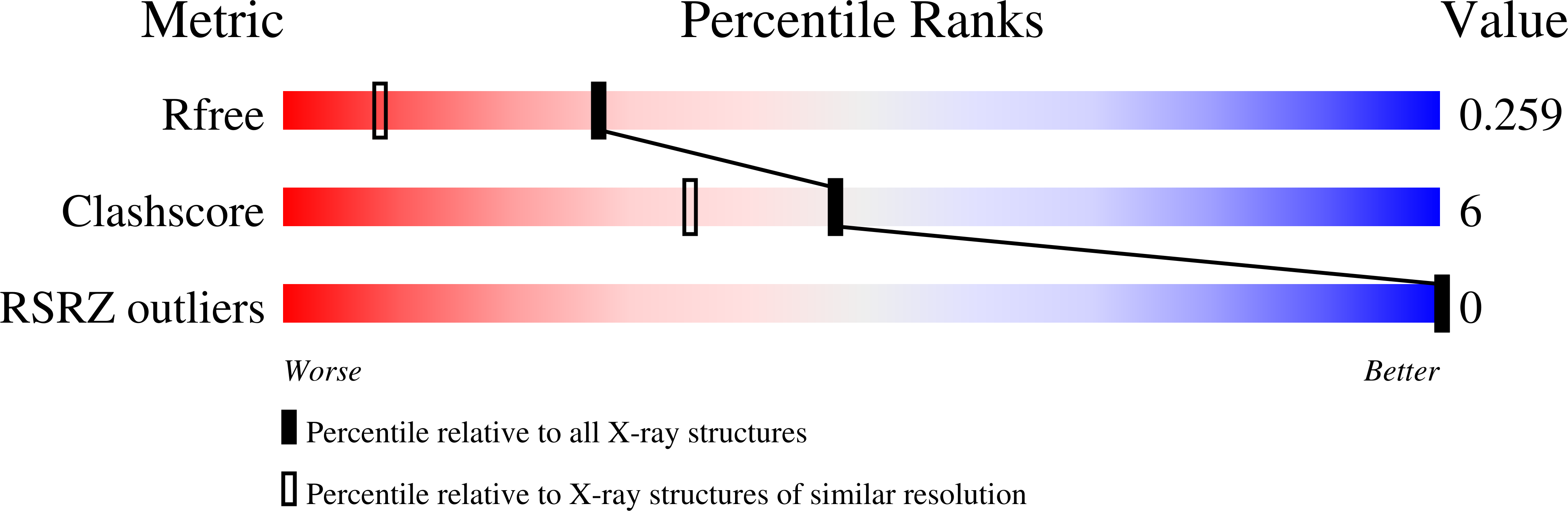Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove
Nguyen, B., Lee, M.P.H., Hamelberg, D., Joubert, A., Bailly, C., Brun, R., Neidle, S., Wilson, W.D.(2002) J Am Chem Soc 124: 13680-13681
- PubMed: 12431090
- DOI: https://doi.org/10.1021/ja027953c
- Primary Citation of Related Structures:
1M6F - PubMed Abstract:
A combination of biophysical techniques has been used to characterize the interaction of an antitrypanosomal agent, CGP 40215A, with DNA. The results from a broad array of methods (DNase I footprinting, surface plasmon resonance, X-ray crystallography, and molecular dynamics) indicate that this compound binds to the minor groove of AT DNA sequences. Despite its unusual linear shape that is not complementary to that of the DNA groove, a high binding affinity was observed in comparison with other similar but more curved diamidine compounds. The amidine groups at both ends of the ligand and the -NH groups on the linker are involved in extensive and dynamic H-bonds to the DNA bases. Complementary and consistent results were obtained from both the X-ray and molecular dynamics studies; both of these methods reveal direct and water-mediated H-bonds between the ligand and the DNA.
Organizational Affiliation:
Department of Chemistry, Georgia State University, Atlanta, Georgia 30303, USA.